6XRZ
 
 | | The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Descriptor: | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Authors: | Zhang, K, Zheludev, I, Hagey, R, Wu, M, Haslecker, R, Hou, Y, Kretsch, R, Pintilie, G, Rangan, R, Kladwang, W, Li, S, Pham, E, Souibgui, C, Baric, R, Sheahan, T, Souza, V, Glenn, J, Chiu, W, Das, R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome.
Biorxiv, 2020
|
|
8FCG
 
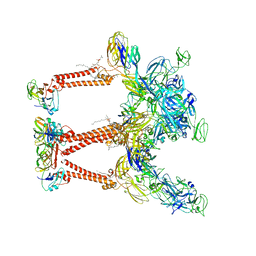 | | Cryo-EM structure of Chikungunya virus asymmetric unit | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Capsid protein, E1 glycoprotein, ... | | Authors: | Su, G.C, Chmielewsk, D, Kaelber, J, Pintilie, G, Chen, M, Jin, J, Auguste, A, Chiu, W. | | Deposit date: | 2022-12-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryogenic electron microscopy and tomography reveal imperfect icosahedral symmetry in alphaviruses.
Pnas Nexus, 3, 2024
|
|
7TNT
 
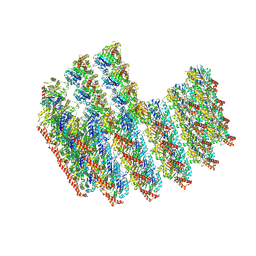 | | The tubulin-based conoid from detergent-extract Toxoplasma gondii cells | | Descriptor: | Tubulin alpha chain, Tubulin beta chain | | Authors: | Sun, S.Y, Pintilie, G.D, Chen, M, Chiu, W. | | Deposit date: | 2022-01-21 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-ET of Toxoplasma parasites gives subnanometer insight into tubulin-based structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TNS
 
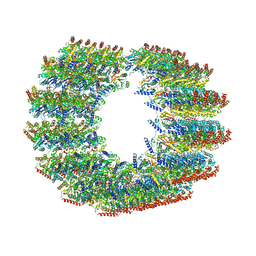 | | Subpellicular microtubule from detergent-extract Toxoplasma gondii cells | | Descriptor: | Microtubule associated protein SPM1, PDI family protein, Tubulin alpha chain, ... | | Authors: | Sun, S.Y, Pintilie, G.D, Chen, M, Chiu, W. | | Deposit date: | 2022-01-21 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryo-ET of Toxoplasma parasites gives subnanometer insight into tubulin-based structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5GAI
 
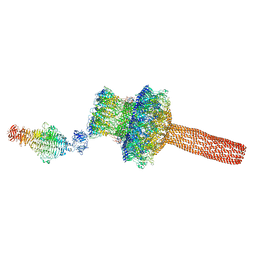 | | Probabilistic Structural Models of Mature P22 Bacteriophage Portal, Hub, and Tailspike proteins | | Descriptor: | Peptidoglycan hydrolase gp4, Portal protein, Tail fiber protein | | Authors: | Pintilie, G, Chen, D.H, Haase-Pettingell, C.A, King, J.A, Chiu, W. | | Deposit date: | 2015-12-01 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Resolution and Probabilistic Models of Components in CryoEM Maps of Mature P22 Bacteriophage.
Biophys.J., 110, 2016
|
|
6DJY
 
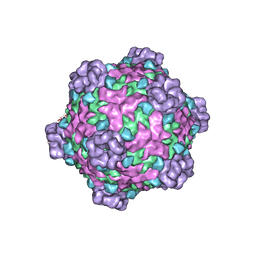 | | Fako virus | | Descriptor: | Clamp protein, Major capsid protein, Turret protein | | Authors: | Kaelber, J.T, Jiang, W, Weaver, S.C, Auguste, A.J, Chiu, W. | | Deposit date: | 2018-05-27 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The polymerase organization of Fako virus
To be Published
|
|
3KTT
 
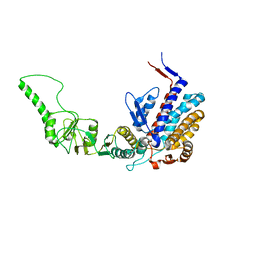 | | Atomic model of bovine TRiC CCT2(beta) subunit derived from a 4.0 Angstrom cryo-EM map | | Descriptor: | T-complex protein 1 subunit beta | | Authors: | Cong, Y, Baker, M.L, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2009-11-25 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7R9H
 
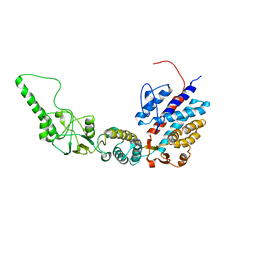 | | Methanococcus maripaludis chaperonin, open conformation 2 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9M
 
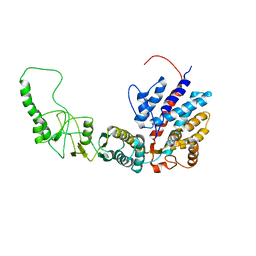 | | Methanococcus maripaludis chaperonin, closed conformation 2 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9J
 
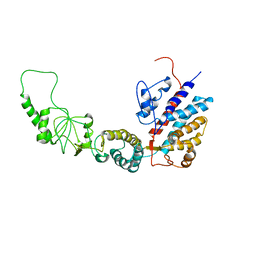 | | Methanococcus maripaludis chaperonin, open conformation 4 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9I
 
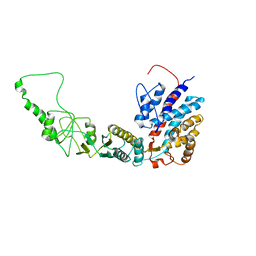 | | Methanococcus maripaludis chaperonin, open conformation 2 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9E
 
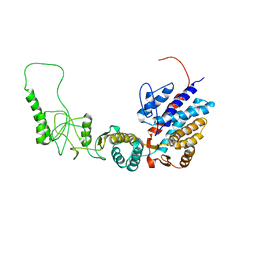 | | Methanococcus maripaludis chaperonin, open conformation 1 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9O
 
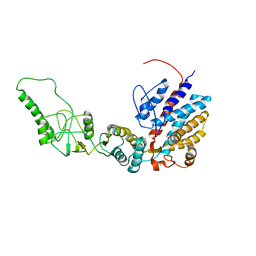 | | Methanococcus maripaludis chaperonin, closed conformation 1 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9K
 
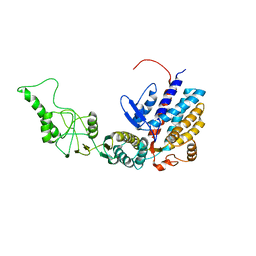 | | Methanococcus maripaludis chaperonin, closed conformation 4 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9U
 
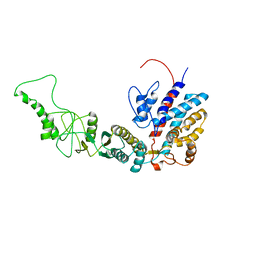 | | Methanococcus maripaludis chaperonin, closed conformation 3 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
8UYJ
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYS
 
 | | SARS-CoV-2 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-2 RNA SL5 domain. | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYL
 
 | | MERS 5' proximal stem-loop 5, conformation 2 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYM
 
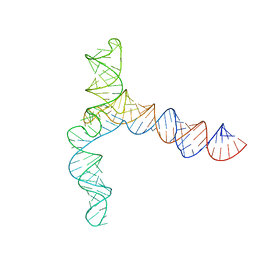 | | MERS 5' proximal stem-loop 5, conformation 3 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYK
 
 | | MERS 5' proximal stem-loop 5, conformation 1 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYE
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 1 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYG
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 2 | | Descriptor: | RNA (135-MER) | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYP
 
 | | SARS-CoV-1 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-1 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6C6L
 
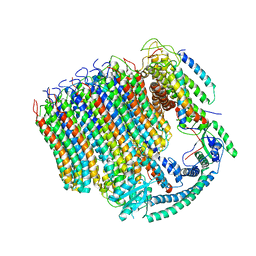 | | Yeast Vacuolar ATPase Vo in lipid nanodisc | | Descriptor: | V-type proton ATPase subunit a, vacuolar isoform, V-type proton ATPase subunit c, ... | | Authors: | Roh, S, Stam, N.J, Hryc, C, Couoh-Cardel, S, Pintilie, G, Chiu, W, Wilkens, S. | | Deposit date: | 2018-01-19 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The 3.5- angstrom CryoEM Structure of Nanodisc-Reconstituted Yeast Vacuolar ATPase VoProton Channel.
Mol. Cell, 69, 2018
|
|
2XYY
 
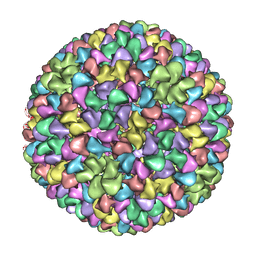 | | De Novo model of Bacteriophage P22 procapsid coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
