1TT9
 
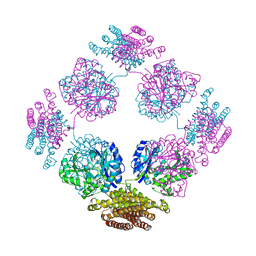 | | Structure of the bifunctional and Golgi associated formiminotransferase cyclodeaminase octamer | | Descriptor: | Formimidoyltransferase-cyclodeaminase (Formiminotransferase- cyclodeaminase) (FTCD) (58 kDa microtubule-binding protein) | | Authors: | Mao, Y, Vyas, N.K, Vyas, M.N, Chen, D.H, Ludtke, S.J, Chiu, W, Quiocho, F.A. | | Deposit date: | 2004-06-22 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structure of the bifunctional and Golgi-associated formiminotransferase cyclodeaminase octamer
Embo J., 23, 2004
|
|
2YEW
 
 | | Modeling Barmah Forest virus structural proteins | | Descriptor: | CAPSID PROTEIN, E1 ENVELOPE GLYCOPROTEIN, E2 ENVELOPE GLYCOPROTEIN | | Authors: | Kostyuchenko, V.A, Jakana, J, Liu, X, Haddow, A.D, Aung, M, Weaver, S.C, Chiu, W, Lok, S.M. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | The Structure of Barmah Forest Virus as Revealed by Cryo-Electron Microscopy at a 6-Angstrom Resolution Has Detailed Transmembrane Protein Architecture and Interactions.
J.Virol., 85, 2011
|
|
3B63
 
 | | Actin filament model in the extended form of acromsomal bundle in the Limulus sperm | | Descriptor: | Actin | | Authors: | Cong, Y, Topf, M, Sali, A, Matsudaira, P, Dougherty, M, Chiu, W, Schmid, M.F. | | Deposit date: | 2007-10-26 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Crystallographic conformers of actin in a biologically active bundle of filaments.
J.Mol.Biol., 375, 2008
|
|
3B5U
 
 | | Actin filament model from extended form of acromsomal bundle in the Limulus sperm | | Descriptor: | Actin, alpha skeletal muscle | | Authors: | Cong, Y, Topf, M, Sali, A, Matsudaira, P, Dougherty, M, Chiu, W, Schmid, M.F. | | Deposit date: | 2007-10-26 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (9.5 Å) | | Cite: | Crystallographic conformers of actin in a biologically active bundle of filaments
J.Mol.Biol., 375, 2008
|
|
5V5S
 
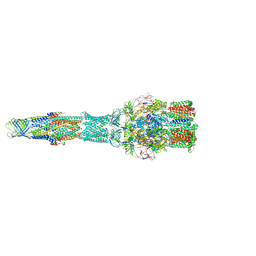 | | multi-drug efflux; membrane transport; RND superfamily; Drug resistance | | Descriptor: | Multidrug efflux pump subunit AcrA, Multidrug efflux pump subunit AcrB, Outer membrane protein TolC | | Authors: | wang, Z, fan, G, Hryc, C.F, Blaza, J.N, Serysheva, I.I, Schmid, M.F, Chiu, W, Luisi, B.F, Du, D. | | Deposit date: | 2017-03-15 | | Release date: | 2017-04-19 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | An allosteric transport mechanism for the AcrAB-TolC Multidrug Efflux Pump.
Elife, 6, 2017
|
|
7K3V
 
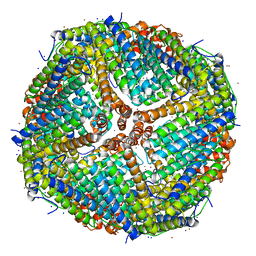 | | Apoferritin structure at 1.34 angstrom resolution determined from a 300 kV Titan Krios G3i electron microscope with K3 detector | | Descriptor: | Ferritin heavy chain, SODIUM ION, ZINC ION | | Authors: | Zhang, K, Pintilie, G, Li, S, Schmid, M, Chiu, W. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.34 Å) | | Cite: | Resolving individual atoms of protein complex by cryo-electron microscopy.
Cell Res., 30, 2020
|
|
7K3W
 
 | | Apoferritin structure at 1.36 angstrom resolution determined from a 300 kV Titan Krios G3i electron microscope with Falcon4 detector | | Descriptor: | Ferritin heavy chain, SODIUM ION, ZINC ION | | Authors: | Zhang, K, Pintilie, G, Li, S, Schmid, M, Chiu, W. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.36 Å) | | Cite: | Resolving individual atoms of protein complex by cryo-electron microscopy.
Cell Res., 30, 2020
|
|
1NO7
 
 | | Structure of the Large Protease Resistant Upper Domain of VP5, the Major Capsid Protein of Herpes Simplex Virus-1 | | Descriptor: | Major capsid protein | | Authors: | Bowman, B.R, Baker, M.L, Rixon, F.J, Chiu, W, Quiocho, F.A. | | Deposit date: | 2003-01-15 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the herpesvirus major capsid protein
Embo J., 22, 2003
|
|
3C9V
 
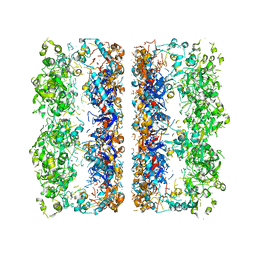 | | C7 Symmetrized Structure of Unliganded GroEL at 4.7 Angstrom Resolution from CryoEM | | Descriptor: | 60 kDa chaperonin | | Authors: | Ludtke, S.J, Baker, M.L, Chen, D.H, Song, J.L, Chuang, D, Chiu, W. | | Deposit date: | 2008-02-18 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | De Novo Backbone Trace of GroEL from Single Particle Electron Cryomicroscopy.
Structure, 16, 2008
|
|
3CAU
 
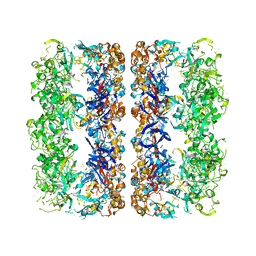 | | D7 symmetrized structure of unliganded GroEL at 4.2 Angstrom resolution by cryoEM | | Descriptor: | 60 kDa chaperonin | | Authors: | Ludtke, S.J, Baker, M.L, Chen, D.H, Song, J.L, Chuang, D, Chiu, W. | | Deposit date: | 2008-02-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | De Novo Backbone Trace of GroEL from Single Particle Electron Cryomicroscopy.
Structure, 16, 2008
|
|
1XPY
 
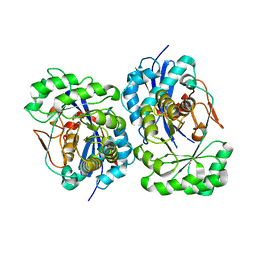 | | Structural Basis for Catalytic Racemization and Substrate Specificity of an N-Acylamino Acid Racemase Homologue from Deinococcus radiodurans | | Descriptor: | MAGNESIUM ION, N-acylamino acid racemase, N~2~-ACETYL-L-GLUTAMINE | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2004-10-10 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
7LUP
 
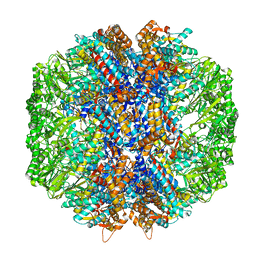 | | Human TRiC/CCT complex with reovirus outer capsid protein sigma-3 | | Descriptor: | Outer capsid protein sigma-3, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Knowlton, J.J, Gestaut, D, Ma, B, Taylor, G, Seven, A.B, Leitner, A, Wilson, G.J, Shanker, S, Yates, N.A, Prasad, B.V.V, Aebersold, R, Chiu, W, Frydman, J, Dermody, T.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural and functional dissection of reovirus capsid folding and assembly by the prefoldin-TRiC/CCT chaperone network.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1XS2
 
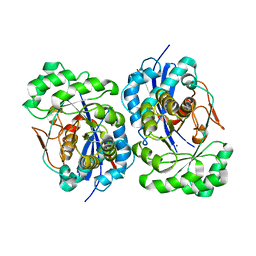 | | Structural Basis for Catalytic Racemization and Substrate Specificity of an N-Acylamino Acid Racemase Homologue from Deinococcus radiodurans | | Descriptor: | MAGNESIUM ION, N-Acylamino Acid Racemase | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
2M5S
 
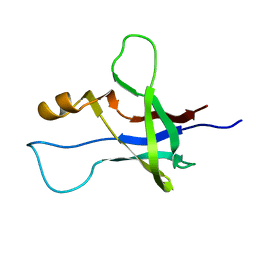 | | High-resolution NMR structure and cryo-EM imaging support multiple functional roles for the accessory I-domain of phage P22 coat protein | | Descriptor: | Coat protein | | Authors: | Rizzo, A.A, Suhanovsky, M.M, Baker, M.L, Fraser, L.C.R, Jones, L.M, Rempel, D.L, Gross, M.L, Chiu, W, Alexandrescu, A.T, Teschke, C.M. | | Deposit date: | 2013-03-05 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Multiple Functional Roles of the Accessory I-Domain of Bacteriophage P22 Coat Protein Revealed by NMR Structure and CryoEM Modeling.
Structure, 22, 2014
|
|
7KIP
 
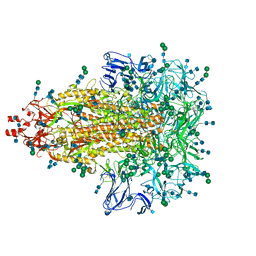 | | A 3.4 Angstrom cryo-EM structure of the human coronavirus spike trimer computationally derived from vitrified NL63 virus particles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Chmielewski, D, Schmid, M, Simmons, G, Jin, J, Chiu, W. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | A 3.4- angstrom cryo-EM structure of the human coronavirus spike trimer computationally derived from vitrified NL63 virus particles.
Biorxiv, 2020
|
|
2L1F
 
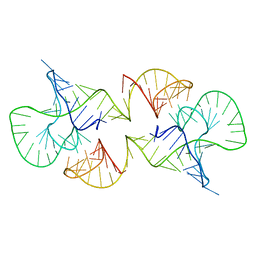 | | Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography | | Descriptor: | RNA (65-MER), RNA (66-MER) | | Authors: | Summers, M.F, Irobalieva, R.N, Tolbert, B, Smalls-Manty, A, Iyalla, K, Loeliger, K, D'Souza, V, Khant, H, Schmid, M, Garcia, E, Telesnitsky, A, Chiu, W, Miyazaki, Y. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography.
J.Mol.Biol., 404, 2010
|
|
6NUD
 
 | | Small conformation of ssRNA-bound CRISPR_Csm complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms protein Csm2, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Zhang, K, Pintilie, G, Li, S, Zhu, Y, Chiu, W, Huang, Z. | | Deposit date: | 2019-01-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Coupling of ssRNA cleavage with DNase activity in type III-A CRISPR-Csm revealed by cryo-EM and biochemistry.
Cell Res., 29, 2019
|
|
6NRB
 
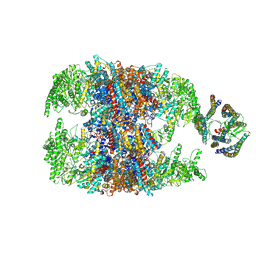 | | hTRiC-hPFD Class2 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6NUE
 
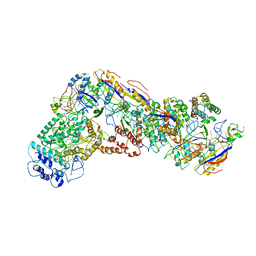 | | Small conformation of apo CRISPR_Csm complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms protein Csm2, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Zhang, K, Pintilie, G, Li, S, Zhu, Y, Chiu, W, Huang, Z. | | Deposit date: | 2019-01-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Coupling of ssRNA cleavage with DNase activity in type III-A CRISPR-Csm revealed by cryo-EM and biochemistry.
Cell Res., 29, 2019
|
|
6NRA
 
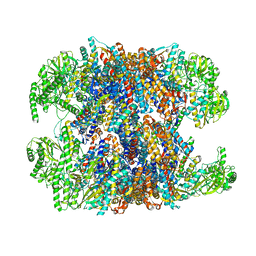 | | hTRiC-hPFD Class1 (No PFD) | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6NRC
 
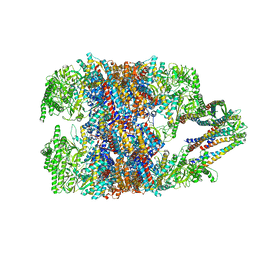 | | hTRiC-hPFD Class3 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6NR9
 
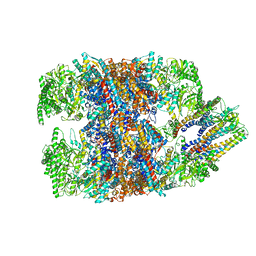 | | hTRiC-hPFD Class5 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6NRD
 
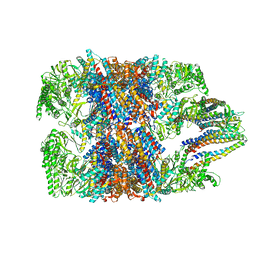 | | hTRiC-hPFD Class4 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6NR8
 
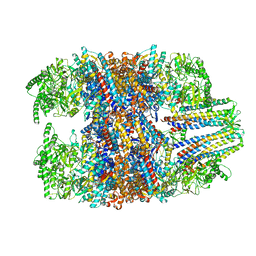 | | hTRiC-hPFD Class6 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6OJN
 
 | | Comparative Model of SGIV Major Coat Protein (MCP) Trimer Based on Cryo-EM Map | | Descriptor: | Major capsid protein | | Authors: | Pintilie, G, Chen, D.-H, Tran, B.N, Jakana, J, Wu, J, Hew, C.L, Chiu, W. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Segmentation and Comparative Modeling in an 8.6- angstrom Cryo-EM Map of the Singapore Grouper Iridovirus.
Structure, 27, 2019
|
|
