1RDV
 
 | |
5H09
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-ethyl2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanoate | | Descriptor: | Tyrosine-protein kinase HCK, ethyl (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-4-methyl-pentanoate | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0B
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanoic acid | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]azaniumyl]-4-methyl-pentanoate, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0H
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-N,N,4-trimethylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-~{N},~{N},4-trimethyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0E
 
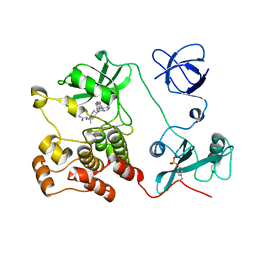 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-4-methyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0G
 
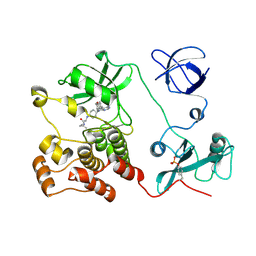 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-N,4-dimethylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-~{N},4-dimethyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
1WUP
 
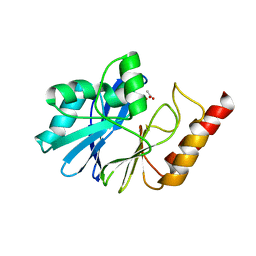 | | Crystal structure of metallo-beta-lactamase IMP-1 mutant (D81E) | | Descriptor: | ACETIC ACID, Beta-lactamase IMP-1, ZINC ION | | Authors: | Yamaguchi, Y, Yamagata, Y, Goto, M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the role of Asp-120(81) of metallo-beta-lactamase (IMP-1) by site-directed mutagenesis, kinetic studies, and X-ray crystallography.
J.Biol.Chem., 280, 2005
|
|
2EWI
 
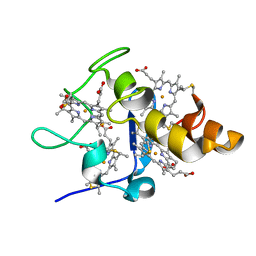 | |
2EWU
 
 | |
1WUO
 
 | | Crystal structure of metallo-beta-lactamase IMP-1 mutant (D81A) | | Descriptor: | ACETIC ACID, Beta-lactamase IMP-1, ZINC ION | | Authors: | Yamaguchi, Y, Yamagata, Y, Goto, M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Probing the role of Asp-120(81) of metallo-beta-lactamase (IMP-1) by site-directed mutagenesis, kinetic studies, and X-ray crystallography.
J.Biol.Chem., 280, 2005
|
|
7C03
 
 | | Crystal structure of POLArISact(T57S), genetically encoded probe for fluorescent polarization | | Descriptor: | POLArISact(T57S) | | Authors: | Tomabechi, Y, Sakai, N, Shirouzu, M. | | Deposit date: | 2020-04-30 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | POLArIS, a versatile probe for molecular orientation, revealed actin filaments associated with microtubule asters in early embryos.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2FMY
 
 | |
3VXD
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N from Streptcoccus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1889, SULFATE ION | | Authors: | Nakamichi, Y, Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2012-09-11 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N from Streptcoccus agalactiae
To be Published
|
|
7EG2
 
 | | Crystal structure of the apoAequorin complex with (S)-daCTZ | | Descriptor: | (2~{S})-2-(hydroxymethyl)-6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-4-(phenylmethyl)-3~{H}-inden-1-one, Aequorin-2 | | Authors: | Tomabechi, Y, Shirouzu, M. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Plos One, 16, 2021
|
|
7EG3
 
 | | Crystal structure of the apoAequorin complex with (S)-HM-daCTZ | | Descriptor: | (2~{S})-6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-4-(phenylmethyl)-2,3-dihydroinden-1-one, Aequorin-2 | | Authors: | Tomabechi, Y, Shirouzu, M. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Plos One, 16, 2021
|
|
1V47
 
 | | Crystal structure of ATP sulfurylase from Thermus thermophillus HB8 in complex with APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, ATP sulfurylase, CHLORIDE ION, ... | | Authors: | Taguchi, Y, Sugishima, M, Fukuyama, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of a novel zinc-binding ATP sulfurylase from Thermus thermophilus HB8
Biochemistry, 43, 2004
|
|
7E9S
 
 | | Archaeal oligosaccharyltransferase AglB from Archaeoglobus fulgidus in complex with an inhibitory peptide and a dolichol-phosphate | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL (7Z)-TETRADEC-7-ENOATE, DI(HYDROXYETHYL)ETHER, Dolichyl-phosphooligosaccharide-protein glycotransferase 3, ... | | Authors: | Taguchi, Y, Hirata, K, Kohda, D. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of an archaeal oligosaccharyltransferase provides insight into the strict exclusion of proline from the N-glycosylation sequon.
Commun Biol, 4, 2021
|
|
6K31
 
 | |
5GV0
 
 | | Crystal structure of the membrane-proximal domain of mouse lysosome-associated membrane protein 1 (LAMP-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome-associated membrane glycoprotein 1, SULFATE ION | | Authors: | Tomabechi, Y, Ehara, H, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-09-01 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2) assemble via distinct modes
Biochem.Biophys.Res.Commun., 479, 2016
|
|
5GV3
 
 | | Crystal structure of the membrane-distal domain of mouse lysosome-associated membrane protein 2 (LAMP-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome-associated membrane glycoprotein 2, ZINC ION | | Authors: | Tomabechi, Y, Ehara, H, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-09-01 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2) assemble via distinct modes.
Biochem. Biophys. Res. Commun., 479, 2016
|
|
3WUX
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae | | Descriptor: | 1,2-ETHANEDIOL, Unsaturated chondroitin disaccharide hydrolase | | Authors: | Nakamichi, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-08 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae
to be published
|
|
3WIW
 
 | | Crystal structure of unsaturated glucuronyl hydrolase specific for heparin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycosyl hydrolase family 88 | | Authors: | Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-26 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a bacterial unsaturated glucuronyl hydrolase with specificity for heparin.
J.Biol.Chem., 289, 2014
|
|
3WVX
 
 | |
3WVY
 
 | | Structure of D48A hen egg white lysozyme in complex with (GlcNAc)4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme C | | Authors: | Kawaguchi, Y, Yoneda, K, Araki, T. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The role of Asp48 in the hydrogen bonding network involving Asp52 of hen egg white lysozyme
TO BE PUBLISHED
|
|
2M32
 
 | | Alpha-1 integrin I-domain in complex with GLOGEN triple helical peptide | | Descriptor: | GLOGEN peptide, Integrin alpha-1, MAGNESIUM ION | | Authors: | Chin, Y, Headey, S, Mohanty, B, McEwan, P, Swarbrick, J, Mulhern, T, Emsley, J, Simpson, J, Scanlon, M. | | Deposit date: | 2013-01-07 | | Release date: | 2013-11-06 | | Last modified: | 2014-02-12 | | Method: | SOLUTION NMR | | Cite: | The Structure of Integrin alpha 1I Domain in Complex with a Collagen-mimetic Peptide.
J.Biol.Chem., 288, 2013
|
|
