2YXW
 
 | | The deletion mutant of Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Higuchi, Y, Komori, H. | | Deposit date: | 2007-04-27 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site
J.Mol.Biol., 373, 2007
|
|
2YXC
 
 | |
2YYW
 
 | |
2CDV
 
 | | REFINED STRUCTURE OF CYTOCHROME C3 AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Higuchi, Y, Kusunoki, M, Matsuura, Y, Yasuoka, N, Kakudo, M. | | Deposit date: | 1983-11-15 | | Release date: | 1984-02-02 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined structure of cytochrome c3 at 1.8 A resolution
J.Mol.Biol., 172, 1984
|
|
2ZWP
 
 | | Crystal structure of Ca3 site mutant of Pro-S324A | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Takeuchi, Y, Tanaka, S, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2008-12-17 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Requirement of a unique Ca(2+)-binding loop for folding of Tk-subtilisin from a hyperthermophilic archaeon.
Biochemistry, 48, 2009
|
|
2YYX
 
 | |
2ZWO
 
 | | Crystal structure of Ca2 site mutant of Pro-S324A | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Takeuchi, Y, Tanaka, S, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2008-12-17 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Requirement of a unique Ca(2+)-binding loop for folding of Tk-subtilisin from a hyperthermophilic archaeon.
Biochemistry, 48, 2009
|
|
2Z47
 
 | |
2ZJD
 
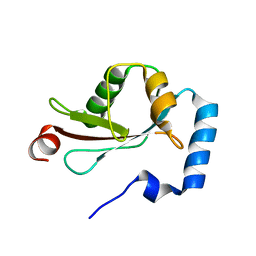 | | Crystal Structure of LC3-p62 complex | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B precursor, undecameric peptide from Sequestosome-1 | | Authors: | Ichimura, Y, Kumanomidou, T, Sou, Y, Mizushima, T, Ezaki, J, Ueno, T, Kominami, E, Yamane, T, Tanaka, K, Komatsu, M. | | Deposit date: | 2008-03-05 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Basis for Sorting Mechanism of p62 in Selective Autophagy
J.Biol.Chem., 283, 2008
|
|
3ANJ
 
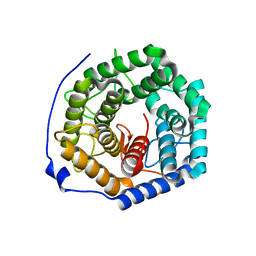 | | Crystal structure of unsaturated glucuronyl hydrolase from Streptcoccus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1889 | | Authors: | Nakamichi, Y, Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural determinants in streptococcal unsaturated glucuronyl hydrolase for recognition of glycosaminoglycan sulfate groups
J.Biol.Chem., 286, 2011
|
|
3ANI
 
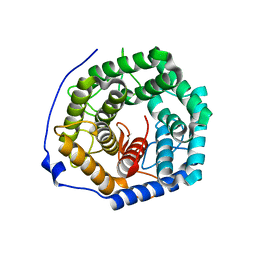 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D175N from Streptcoccus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1889 | | Authors: | Nakamichi, Y, Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural determinants in streptococcal unsaturated glucuronyl hydrolase for recognition of glycosaminoglycan sulfate groups
J.Biol.Chem., 286, 2011
|
|
3ANK
 
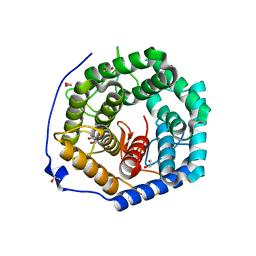 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D175N from Streptcoccus agalactiae complexed with dGlcA-GalNAc6S | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, Putative uncharacterized protein gbs1889 | | Authors: | Nakamichi, Y, Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural determinants in streptococcal unsaturated glucuronyl hydrolase for recognition of glycosaminoglycan sulfate groups
J.Biol.Chem., 286, 2011
|
|
7JPM
 
 | |
3AQO
 
 | |
2FFN
 
 | |
2EWK
 
 | |
2ZJW
 
 | | Crystal structure of human CK2 alpha complexed with Ellagic acid | | Descriptor: | 2,3,7,8-tetrahydroxychromeno[5,4,3-cde]chromene-5,10-dione, Casein kinase II subunit alpha | | Authors: | Sekiguchi, Y, Kinoshita, T, Nakaniwa, T, Tada, T. | | Deposit date: | 2008-03-11 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into human CK2alpha in complex with the potent inhibitor ellagic acid
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2EWI
 
 | |
2EWU
 
 | |
3AMY
 
 | | Crystal structure of human CK2 alpha complexed with apigenin | | Descriptor: | 1,2-ETHANEDIOL, 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Casein kinase II subunit alpha | | Authors: | Sekiguchi, Y, Nakaniwa, T, Kinoshita, T, Tada, T. | | Deposit date: | 2010-08-25 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human CK2 alpha complexed with apigenin
To be Published
|
|
3UYV
 
 | | Crystal structure of a glycosylated ice-binding protein (LeIBP) from Arctic yeast | | Descriptor: | Antifreeze protein, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lee, J.H, Park, A.K, Do, H, Park, K.S, Moh, S.H, Chi, Y.M, Kim, H.J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for the antifreeze activity of an ice-binding protein from an Arctic yeast.
J.Biol.Chem., 2012
|
|
2G04
 
 | | Crystal structure of fatty acid-CoA racemase from Mycobacterium tuberculosis H37Rv | | Descriptor: | PROBABLE FATTY-ACID-CoA RACEMASE FAR | | Authors: | Lee, K.S, Park, S.M, Rhee, K.H, Bang, W.G, Hwang, K.Y, Chi, Y.M. | | Deposit date: | 2006-02-11 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of fatty acid-CoA racemase from Mycobacterium tuberculosis H37Rv
Proteins, 64, 2006
|
|
6OH1
 
 | | IgA1 Protease G5 domain structure | | Descriptor: | Immunoglobulin A1 protease | | Authors: | Eisenmesser, E.Z, Chi, Y.C, Paukovich, N, Redzic, J.S, Rahkola, J.T, Janoff, E.N. | | Deposit date: | 2019-04-04 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Streptococcus pneumoniae G5 domains bind different ligands.
Protein Sci., 28, 2019
|
|
4YWU
 
 | | Structural insight into the substrate inhibition mechanism of NADP+-dependent succinic semialdehyde dehydrogenase from Streptococcus pyogenes | | Descriptor: | 4-oxobutanoic acid, SULFATE ION, Succinic semialdehyde dehydrogenase | | Authors: | Jang, E.H, Park, S.A, Chi, Y.M, Lee, K.S. | | Deposit date: | 2015-03-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the substrate inhibition mechanism of NADP(+)-dependent succinic semialdehyde dehydrogenase from Streptococcus pyogenes.
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YWV
 
 | | Structural insight into the substrate inhibition mechanism of NADP+-dependent succinic semialdehyde dehydrogenase from Streptococcus pyogenes | | Descriptor: | 4-oxobutanoic acid, SULFATE ION, Succinic semialdehyde dehydrogenase | | Authors: | Park, S.A, Jang, E.H, Chi, Y.M, Lee, K.S. | | Deposit date: | 2015-03-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the substrate inhibition mechanism of NADP(+)-dependent succinic semialdehyde dehydrogenase from Streptococcus pyogenes.
Biochem.Biophys.Res.Commun., 461, 2015
|
|
