3ATE
 
 | |
2D5M
 
 | |
2DOO
 
 | | The structure of IMP-1 complexed with the detecting reagent (DansylC4SH) by a fluorescent probe | | Descriptor: | BETA-LACTAMASE IMP-1, N-[4-({[5-(DIMETHYLAMINO)-1-NAPHTHYL]SULFONYL}AMINO)BUTYL]-3-SULFANYLPROPANAMIDE, ZINC ION | | Authors: | Kurosaki, H, Yamaguchi, Y, Yasuzawa, H, Jin, W, Yamagata, Y, Arakawa, Y. | | Deposit date: | 2006-05-01 | | Release date: | 2006-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Probing, inhibition, and crystallographic characterization of metallo-beta-lactamase (IMP-1) with fluorescent agents containing dansyl and thiol groups
Chemmedchem, 1, 2006
|
|
2E4F
 
 | |
2E4I
 
 | | Human Telomeric DNA mixed-parallel/antiparallel quadruplex under Physiological Ionic Conditions Stabilized by Proper Incorporation of 8-Bromoguanosines | | Descriptor: | DNA (5'-D(*DAP*(BGM)P*DGP*DGP*DTP*DTP*DAP*(BGM)P*DGP*DGP*DTP*DTP*DAP*(BGM)P*(BGM)P*DGP*DTP*DTP*DAP*(BGM)P*DGP*DG)-3') | | Authors: | Matsugami, A, Xu, Y, Noguchi, Y, Sugiyama, H, Katahira, M. | | Deposit date: | 2006-12-11 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K+ solution
Febs J., 274, 2007
|
|
2DCF
 
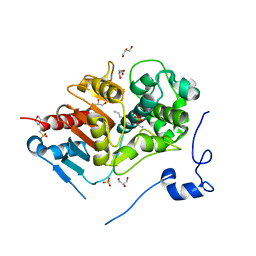 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N mutant with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2006-01-06 | | Release date: | 2007-01-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nylon-oligomer degrading enzyme/substrate complex: catalytic mechanism of 6-aminohexanoate-dimer hydrolase
J.Mol.Biol., 370, 2007
|
|
2ZC4
 
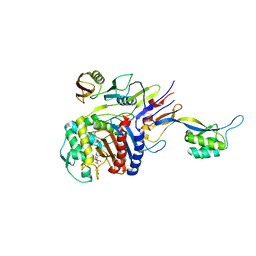 | | Penicillin-binding protein 2X (PBP 2X) acyl-enzyme complex (tebipenem) from Streptococcus pneumoniae | | Descriptor: | (4R,5S)-3-(1-(4,5-dihydrothiazol-2-yl)azetidin-3-ylthio)-5-((2S,3R)-3-hydroxy-1-oxobutan-2-yl)-4-methyl-4,5- dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 2X, SULFATE ION | | Authors: | Yamada, M, Watanabe, T, Takeuchi, Y. | | Deposit date: | 2007-11-02 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Biapenem and Tebipenem Complexed with Penicillin-Binding Proteins 2X and 1A from Streptococcus pneumoniae
Antimicrob.Agents Chemother., 52, 2008
|
|
3A33
 
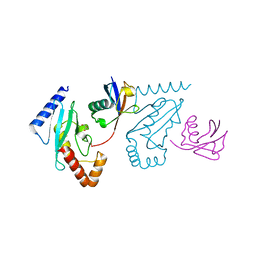 | | UbcH5b~Ubiquitin Conjugate | | Descriptor: | GLYCEROL, Ubiquitin, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Sakata, E, Satoh, T, Yamamoto, S, Yamaguchi, Y, Yagi-Utsumi, M, Kurimoto, E, Wakatsuki, S, Kato, K. | | Deposit date: | 2009-06-08 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of UbcH5b~Ubiquitin Intermediate: Insight into the Formation of the Self-Assembled E2~Ub Conjugates
Structure, 18, 2010
|
|
2ZCB
 
 | | Crystal Structure of ubiquitin P37A/P38A | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Kitahara, R, Tanaka, T, Sakata, E, Yamaguchi, Y, Kato, K, Yokoyama, S. | | Deposit date: | 2007-11-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of ubiquitin P37A/P38A
To be published
|
|
2ZC3
 
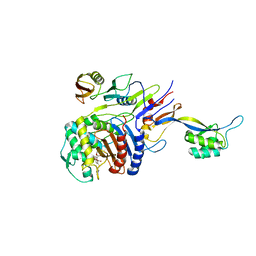 | | Penicillin-binding protein 2X (PBP 2X) acyl-enzyme complex (biapenem) from Streptococcus pneumoniae | | Descriptor: | (4R,5S)-3-(6,7-dihydro-5H-pyrazolo[1,2-a][1,2,4]triazol-4-ium-6-ylsulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-meth yl-4,5-dihydro-1H-pyrrole-2-carboxylate, Penicillin-binding protein 2X, SULFATE ION | | Authors: | Yamada, M, Watanabe, T, Takeuchi, Y. | | Deposit date: | 2007-11-02 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Biapenem and Tebipenem Complexed with Penicillin-Binding Proteins 2X and 1A from Streptococcus pneumoniae
Antimicrob.Agents Chemother., 52, 2008
|
|
5I8N
 
 | |
3AVE
 
 | | Crystal Structure of the Fucosylated Fc Fragment from Human Immunoglobulin G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, ZINC ION | | Authors: | Matsumiya, S, Yamaguchi, Y, Saito, J, Nagano, M, Sasakawa, H, Otaki, S, Satoh, M, Shitara, K, Kato, K. | | Deposit date: | 2011-03-04 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Corrigendum to "Structural Comparison of Fucosylated and Nonfucosylated Fc Fragments of Human Immunoglobulin G1" [J. Mol. Biol. 386/3 (2007) 767-779]
J.Mol.Biol., 408, 2011
|
|
2D5G
 
 | |
2D1P
 
 | | crystal structure of heterohexameric TusBCD proteins, which are crucial for the tRNA modification | | Descriptor: | Hypothetical UPF0116 protein yheM, Hypothetical UPF0163 protein yheN, Hypothetical protein yheL, ... | | Authors: | Numata, T, Fukai, S, Ikeuchi, Y, Suzuki, T, Nureki, O. | | Deposit date: | 2005-08-30 | | Release date: | 2006-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Sulfur Relay to RNA Mediated by Heterohexameric TusBCD Complex
Structure, 14, 2006
|
|
2DPF
 
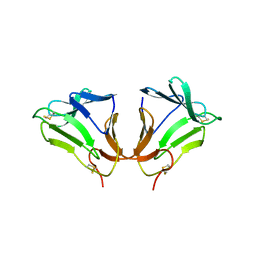 | | Crystal Structure of curculin1 homodimer | | Descriptor: | Curculin, SULFATE ION | | Authors: | Kurimoto, E, Suzuki, M, Amemiya, E, Yamaguchi, Y, Nirasawa, S, Shimba, N, Xu, N, Kashiwagi, T, Kawai, M, Suzuki, E, Kato, K. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Curculin Exhibits Sweet-tasting and Taste-modifying Activities through Its Distinct Molecular Surfaces.
J.Biol.Chem., 282, 2007
|
|
2YYS
 
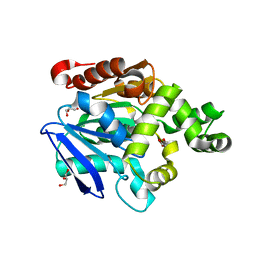 | | Crystal structure of the proline iminopeptidase-related protein TTHA1809 from Thermus thermophilus HB8 | | Descriptor: | GLYCEROL, Proline iminopeptidase-related protein | | Authors: | Okai, M, Miyauchi, Y, Ebihara, A, Lee, W.C, Nagata, K, Tanokura, M. | | Deposit date: | 2007-05-01 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the proline iminopeptidase-related protein TTHA1809 from Thermus thermophilus HB8
Proteins, 70, 2008
|
|
2Z2M
 
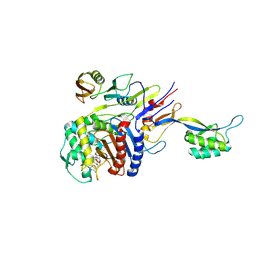 | | Cefditoren-Acylated Penicillin-Binding Protein 2X (PBP2X) from Streptococcus pneumoniae | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(2-AMINO-1,3-THIAZOL-4-YL)-2-(METHOXYIMINO)ACETYL]AMINO}-2-OXOETHYL]-5-[(Z)-2-(4-METHYL-1,3-THIAZOL-5-YL)VINYL]-3,6-DIHYDRO-2H-1,3-THIAZINE-4-CARBOXYLIC ACID, Penicillin-binding protein 2X, SULFATE ION | | Authors: | Yamada, M, Watanabe, T, Takeuchi, Y. | | Deposit date: | 2007-05-23 | | Release date: | 2007-09-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Cefditoren Complexed with Streptococcus pneumoniae Penicillin-Binding Protein 2X: Structural Basis for its High Antimicrobial Activity
Antimicrob.Agents Chemother., 51, 2007
|
|
2DER
 
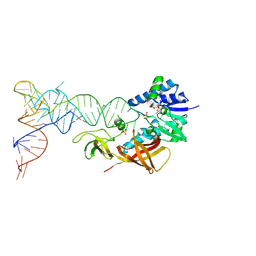 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the initial tRNA binding state | | Descriptor: | PHOSPHATE ION, SULFATE ION, tRNA, ... | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
2DET
 
 | | Cocrystal structure of an RNA sulfuration enzyme MnmA and tRNA-Glu in the pre-reaction state | | Descriptor: | SULFATE ION, tRNA, tRNA-specific 2-thiouridylase mnmA | | Authors: | Numata, T, Ikeuchi, Y, Fukai, S, Suzuki, T, Nureki, O. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of tRNA sulphuration via an adenylated intermediate
Nature, 442, 2006
|
|
2DJJ
 
 | |
2ZPC
 
 | | Crystal structure of the R43L mutant of LolA in the closed form | | Descriptor: | Outer-membrane lipoprotein carrier protein | | Authors: | Takeda, K, Yokota, N, Oguchi, Y, Tokuda, H, Miki, K. | | Deposit date: | 2008-07-10 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Opening and closing of the hydrophobic cavity of LolA coupled to lipoprotein binding and release.
J.Biol.Chem., 283, 2008
|
|
2ZPD
 
 | | Crystal structure of the R43L mutant of LolA in the open form | | Descriptor: | Outer-membrane lipoprotein carrier protein | | Authors: | Takeda, K, Yokota, N, Oguchi, Y, Tokuda, H, Miki, K. | | Deposit date: | 2008-07-10 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Opening and closing of the hydrophobic cavity of LolA coupled to lipoprotein binding and release.
J.Biol.Chem., 283, 2008
|
|
2ZWN
 
 | | Crystal structure of the novel two-domain type laccase from a metagenome | | Descriptor: | CHLORIDE ION, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Komori, H, Miyazaki, K, Higuchi, Y. | | Deposit date: | 2008-12-17 | | Release date: | 2009-04-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of a two-domain type laccase: a missing link in the evolution of multi-copper proteins
Febs Lett., 583, 2009
|
|
2Z2L
 
 | |
2DTS
 
 | | Crystal Structure of the Defucosylated Fc Fragment from Human Immunoglobulin G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Matsumiya, S, Yamaguchi, Y, Saito, J, Nagano, M, Sasakawa, H, Otaki, S, Satoh, M, Shitara, K, Kato, K. | | Deposit date: | 2006-07-14 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural comparison of fucosylated and nonfucosylated fc fragments of human immunoglobulin g1
J.Mol.Biol., 368, 2007
|
|
