3VS4
 
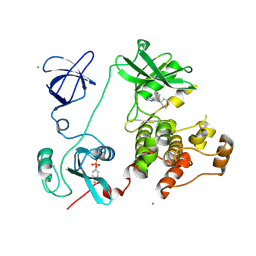 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 5-(4-phenoxyphenyl)-7-(tetrahydro-2H-pyran-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | 分子名称: | 5-(4-phenoxyphenyl)-7-(tetrahydro-2H-pyran-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | 登録日 | 2012-04-21 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.747 Å) | | 主引用文献 | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
6LTM
 
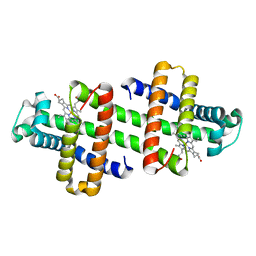 | | The dimeric structure of G80A/H81A/H82A myoglobin | | 分子名称: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | 登録日 | 2020-01-22 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
3VWQ
 
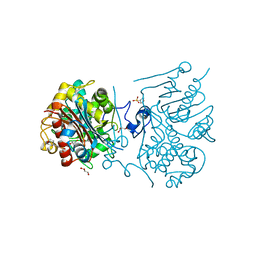 | | 6-aminohexanoate-dimer hydrolase S112A/G181D/R187A/H266N/D370Y mutant complexd with 6-aminohexanoate | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | 著者 | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | 登録日 | 2012-08-30 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VQT
 
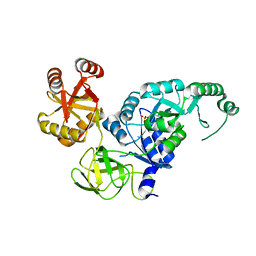 | | Crystal structure analysis of the translation factor RF3 | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Peptide chain release factor 3 | | 著者 | Kihira, K, Shomura, Y, Shibata, N, Kitamura, M, Higuchi, Y. | | 登録日 | 2012-03-30 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure analysis of the translation factor RF3 (release factor 3)
Febs Lett., 586, 2012
|
|
5YOT
 
 | | Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Matsuzawa, T, Watanabe, M, Nakamichi, Y, Yaoi, K. | | 登録日 | 2017-10-31 | | 公開日 | 2018-11-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal structure and substrate recognition mechanism of Aspergillus oryzae isoprimeverose-producing enzyme.
J.Struct.Biol., 205, 2019
|
|
3VS0
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor N-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]benzamide | | 分子名称: | CALCIUM ION, CHLORIDE ION, N-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]benzamide, ... | | 著者 | Kuratani, M, Honda, K, Tomabechi, Y, Handa, N, Yokoyama, S. | | 登録日 | 2012-04-21 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.934 Å) | | 主引用文献 | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
6LTL
 
 | | The dimeric structure of G80A myoglobin | | 分子名称: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | 登録日 | 2020-01-22 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
3VR1
 
 | | Crystal structure analysis of the translation factor RF3 | | 分子名称: | GUANOSINE-5',3'-TETRAPHOSPHATE, Peptide chain release factor 3 | | 著者 | Kihira, K, Shomura, Y, Shibata, N, Kitamura, M, Higuchi, Y. | | 登録日 | 2012-04-03 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure analysis of the translation factor RF3 (release factor 3)
Febs Lett., 586, 2012
|
|
3VS6
 
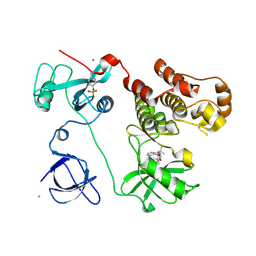 | | Crystal structure of HCK complexed with a pyrazolo-pyrimidine inhibitor tert-butyl {4-[4-amino-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl]-2-methoxyphenyl}carbamate | | 分子名称: | CALCIUM ION, CHLORIDE ION, Tyrosine-protein kinase HCK, ... | | 著者 | Kuratani, M, Honda, K, Tomabechi, Y, Toyama, M, Handa, N, Yokoyama, S. | | 登録日 | 2012-04-21 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.373 Å) | | 主引用文献 | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS3
 
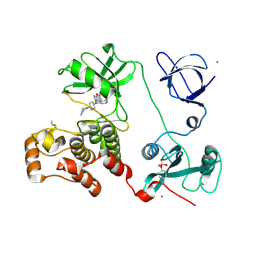 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | 分子名称: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Kuratani, M, Tomaebchi, Y, Handa, N, Yokoyama, S. | | 登録日 | 2012-04-21 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VWP
 
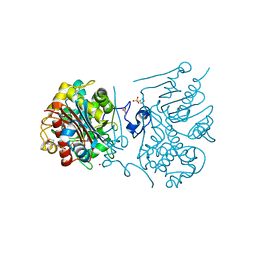 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187S/H266N/D370Y mutant complexd with 6-aminohexanoate | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | 著者 | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | 登録日 | 2012-08-30 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VS5
 
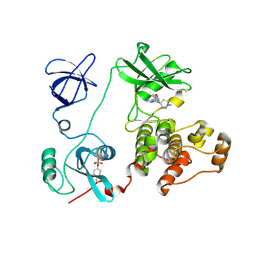 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | 分子名称: | 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, Tyrosine-protein kinase HCK | | 著者 | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | 登録日 | 2012-04-21 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.851 Å) | | 主引用文献 | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VWN
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187G/H266N/D370Y mutant | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | 著者 | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | 登録日 | 2012-08-30 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
2LXZ
 
 | | Solution Structure of the Antimicrobial Peptide Human Defensin 5 | | 分子名称: | Defensin-5 | | 著者 | Wommack, A.J, Robson, S.A, Wanniarahchi, Y.A, Wan, A, Turner, C.J, Nolan, E.M. | | 登録日 | 2012-09-10 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure and condition-dependent oligomerization of the antimicrobial Peptide human defensin 5.
Biochemistry, 51, 2012
|
|
3WII
 
 | | Crystal structure of the Fab fragment of B2212A, a murine monoclonal antibody specific for the third fibronectin domain (Fn3) of human ROBO1. | | 分子名称: | anti-human ROBO1 antibody B2212A Fab heavy chain, anti-human ROBO1 antibody B2212A Fab light chain | | 著者 | Nakayama, T, Mizohata, E, Yamashita, T, Nagatoishi, S, Nakakido, M, Iwanari, H, Mochizuki, Y, Kado, Y, Yokota, Y, Sato, R, Tsumoto, K, Fujitani, H, Kodama, T, Hamakubo, T, Inoue, T. | | 登録日 | 2013-09-12 | | 公開日 | 2015-01-21 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses
Protein Sci., 24, 2015
|
|
3FDF
 
 | | Crystal structure of the serine phosphatase of RNA polymerase II CTD (SSU72 superfamily) from Drosophila melanogaster. Orthorhombic crystal form. Northeast Structural Genomics Consortium target FR253. | | 分子名称: | FR253 | | 著者 | Kuzin, A.P, Chen, Y, Seetharaman, J, Forouhar, F, Chinag, Y, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-11-25 | | 公開日 | 2009-01-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Orthorombic crystal structure of serine phosphatase of rna polymerase ii ctd from fly drosofila melanogaster. northeast structural genomics consortium target fr253.
To be Published
|
|
3WIH
 
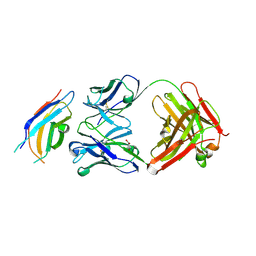 | | Crystal structure of the third fibronectin domain (Fn3) of human ROBO1 in complex with the Fab fragment of murine monoclonal antibody B2212A. | | 分子名称: | GLYCEROL, Roundabout homolog 1, anti-human ROBO1 antibody B2212A Fab heavy chain, ... | | 著者 | Nakayama, T, Mizohata, E, Yamashita, T, Nagatoishi, M, Iwanari, H, Mochizuki, Y, Kado, Y, Yokota, Y, Sato, R, Tsumoto, K, Fujitani, H, Kodama, T, Hamakubo, T, Inoue, T. | | 登録日 | 2013-09-12 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses
Protein Sci., 24, 2015
|
|
3FMV
 
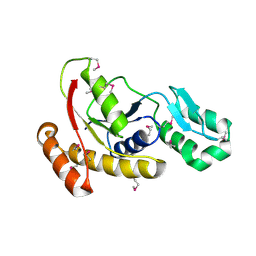 | | Crystal structure of the serine phosphatase of RNA polymerase II CTD (SSU72 superfamily) from Drosophila melanogaster. Monoclinic crystal form. Northeast Structural Genomics Consortium target FR253. | | 分子名称: | Serine phosphatase of RNA polymerase II CTD | | 著者 | Kuzin, A.P, Chen, Y, Seetharaman, J, Forouhar, F, Chinag, Y, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-12-22 | | 公開日 | 2009-01-06 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Crystal structure of the serine phosphatase of RNA polymerase II CTD (SSU72 superfamily) from Drosophila melanogaster. Monoclinic crystal form. Northeast Structural Genomics Consortium target FR253.
To be Published
|
|
3WKV
 
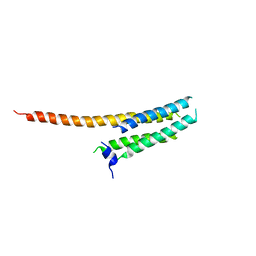 | | Voltage-gated proton channel: VSOP/Hv1 chimeric channel | | 分子名称: | Ion channel | | 著者 | Takeshita, K, Sakata, S, Yamashita, E, Fujiwara, Y, Kawanabe, A, Kurokawa, T, Okochi, Y, Matsuda, M, Narita, H, Okamura, Y, Nakagawa, A. | | 登録日 | 2013-10-31 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.453 Å) | | 主引用文献 | X-ray crystal structure of voltage-gated proton channel.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4MMH
 
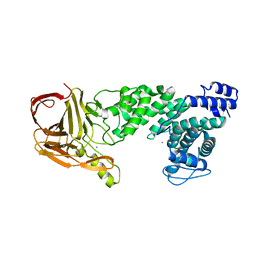 | | Crystal structure of heparan sulfate lyase HepC from Pedobacter heparinus | | 分子名称: | CALCIUM ION, Heparinase III protein | | 著者 | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2013-09-09 | | 公開日 | 2014-01-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
4MMI
 
 | | Crystal structure of heparan sulfate lyase HepC mutant from Pedobacter heparinus | | 分子名称: | CALCIUM ION, Heparinase III protein | | 著者 | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2013-09-09 | | 公開日 | 2014-01-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
1C16
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE GAMMA/DELTA T CELL LIGAND T22 | | 分子名称: | MHC-LIKE PROTEIN T22, PROTEIN (BETA-2-MICROGLOBULIN) | | 著者 | Wingren, C, Crowley, M.P, Degano, M, Chien, Y, Wilson, I.A. | | 登録日 | 1999-07-20 | | 公開日 | 2000-01-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure of a gammadelta T cell receptor ligand T22: a truncated MHC-like fold.
Science, 287, 2000
|
|
7VU6
 
 | | The crystal structure of SARS-CoV-2 3CL protease in complex with compound 3 | | 分子名称: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Yamamoto, S, Yamane, J, Tachibana, Y. | | 登録日 | 2021-11-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19.
J.Med.Chem., 65, 2022
|
|
5YQS
 
 | | Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Yaoi, K, Matsuzawa, T, Watanabe, M, Nakamichi, Y. | | 登録日 | 2017-11-07 | | 公開日 | 2018-11-14 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure and substrate recognition mechanism of Aspergillus oryzae isoprimeverose-producing enzyme.
J.Struct.Biol., 205, 2019
|
|
3T5W
 
 | | 2ME modified human SOD1 | | 分子名称: | COPPER (I) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | 著者 | Ihara, K, Yamaguchi, Y, Torigoe, H, Wakatsuki, S, Taniguchi, N, Suzuki, K, Fujiwara, N. | | 登録日 | 2011-07-28 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural switching of Cu,Zn-superoxide dismutases at loop VI: insights from the crystal structure of 2-mercaptoethanol-modified enzyme
Biosci.Rep., 32, 2012
|
|
