7MTG
 
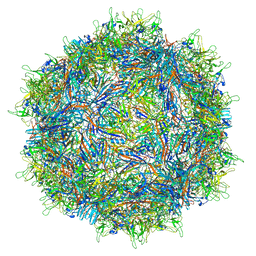 | | Structure of the adeno-associated virus 9 capsid at pH 6.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTP
 
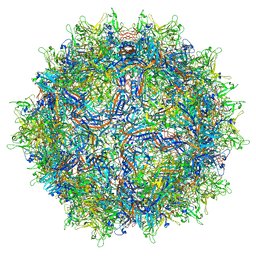 | | Structure of the adeno-associated virus 9 capsid at pH 5.5 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTW
 
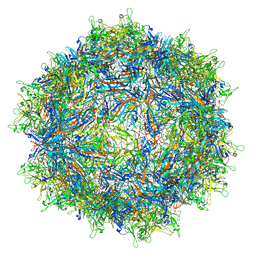 | | Structure of the adeno-associated virus 9 capsid at pH 4.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
1Z8Y
 
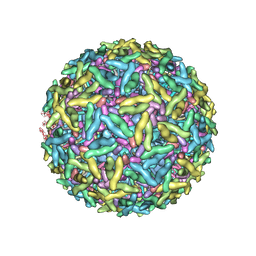 | | Mapping the E2 Glycoprotein of Alphaviruses | | Descriptor: | Capsid protein C, Spike glycoprotein E1, Spike glycoprotein E2 | | Authors: | Mukhopadhyay, S, Zhang, W, Gabler, S, Chipman, P.R, Strauss, E.G, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2005-03-31 | | Release date: | 2006-02-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses.
Structure, 14, 2006
|
|
7MDO
 
 | | Structure of human p97 ATPase L464P mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Zhang, X, Gui, L, Li, S, Nandi, P, Columbres, R.C, Wong, D.E, Moen, D.R, Lin, H.J, Chiu, P.-L, Chou, T.-F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Conserved L464 in p97 D1-D2 linker is critical for p97 cofactor regulated ATPase activity.
Biochem.J., 478, 2021
|
|
7MDM
 
 | | Structure of human p97 ATPase L464P mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Zhang, X, Gui, L, Li, S, Nandi, P, Columbres, R.C, Wong, D.E, Moen, D.R, Lin, H.J, Chiu, P.-L, Chou, T.-F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.86 Å) | | Cite: | Conserved L464 in p97 D1-D2 linker is critical for p97 cofactor regulated ATPase activity.
Biochem.J., 478, 2021
|
|
4JNB
 
 | | Crystal structure of the Catalytic Domain of Human DUSP12 | | Descriptor: | Dual specificity protein phosphatase 12, SULFATE ION | | Authors: | Jeon, T.J, Chien, P.N, Ku, B, Kim, S.J, Ryu, S.E. | | Deposit date: | 2013-03-15 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The family-wide structure and function of human dual-specificity protein phosphatases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CHW
 
 | | The electron crystallography structure of the cAMP-free potassium channel MloK1 | | Descriptor: | CYCLIC NUCLEOTIDE-GATED POTASSIUM CHANNEL MLL3241, POTASSIUM ION | | Authors: | Kowal, J, Chami, M, Baumgartner, P, Arheit, M, Chiu, P.L, Rangl, M, Scheuring, S, Schroeder, G.F, Nimigean, C.M, Stahlberg, H. | | Deposit date: | 2013-12-04 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Ligand-induced structural changes in the cyclic nucleotide-modulated potassium channel MloK1.
Nat Commun, 5, 2014
|
|
1DS6
 
 | | CRYSTAL STRUCTURE OF A RAC-RHOGDI COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAS-RELATED C3 BOTULINUM TOXIN SUBSTRATE 2, ... | | Authors: | Scheffzek, K, Stephan, I, Jensen, O.N, Illenberger, D, Gierschik, P. | | Deposit date: | 2000-01-07 | | Release date: | 2000-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Rac-RhoGDI complex and the structural basis for the regulation of Rho proteins by RhoGDI.
Nat.Struct.Biol., 7, 2000
|
|
1DG5
 
 | | DIHYDROFOLATE REDUCTASE OF MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND TRIMETHOPRIM | | Descriptor: | DIHYDROFOLATE REDUCTASE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, R, Sirawaraporn, R, Chitnumsub, P, Sirawaraporn, W, Wooden, J, Athappilly, F, Turley, S, Hol, W.G. | | Deposit date: | 1999-11-23 | | Release date: | 2000-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs.
J.Mol.Biol., 295, 2000
|
|
1DG8
 
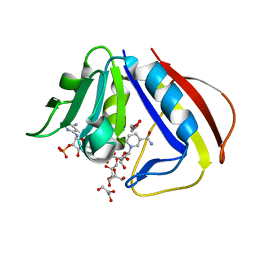 | | DIHYDROFOLATE REDUCTASE OF MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH | | Descriptor: | DIHYDROFOLATE REDUCTASE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, R, Sirawaraporn, R, Chitnumsub, P, Sirawaraporn, W, Wooden, J, Athappilly, F, Turley, S, Hol, W.G. | | Deposit date: | 1999-11-23 | | Release date: | 2000-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs.
J.Mol.Biol., 295, 2000
|
|
2AZX
 
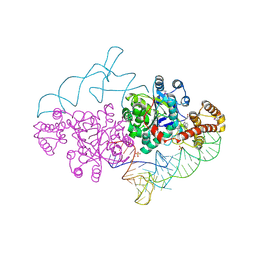 | | Charged and uncharged tRNAs adopt distinct conformations when complexed with human tryptophanyl-tRNA synthetase | | Descriptor: | 72-MER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Yang, X.L, Otero, F.J, Ewalt, K.L, Liu, J, Swairjo, M.A, Kohrer, C, RajBhandary, U.L, Skene, R.J, McRee, D.E, Schimmel, P. | | Deposit date: | 2005-09-12 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis.
Embo J., 25, 2006
|
|
2QZD
 
 | | Fitted structure of SCR4 of DAF into cryoEM density | | Descriptor: | Complement decay-accelerating factor | | Authors: | Hafenstein, S, Bowman, V.D, Chipman, P.R, Bator Kelly, C.M, Lin, F, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2007-08-16 | | Release date: | 2007-10-30 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Interaction of decay-accelerating factor with coxsackievirus b3.
J.Virol., 81, 2007
|
|
7E8P
 
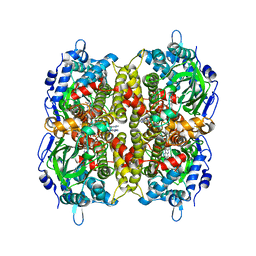 | | Crystal structure of a Flavin-dependent Monooxygenase HadA wild type complexed with reduced FAD and 4-nitrophenol | | Descriptor: | Chlorophenol monooxygenase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, P-NITROPHENOL | | Authors: | Pimviriyakul, P, Jaruwat, A, Chitnumsub, P, Chaiyen, P. | | Deposit date: | 2021-03-02 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into a flavin-dependent dehalogenase HadA explain catalysis and substrate inhibition via quadruple pi-stacking.
J.Biol.Chem., 297, 2021
|
|
7E8Q
 
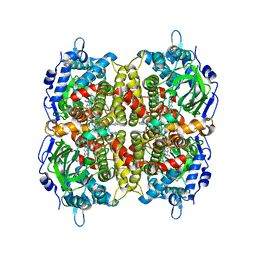 | | Crystal structure of a Flavin-dependent Monooxygenase HadA F441V mutant complexed with reduced FAD and 4-nitrophenol | | Descriptor: | Chlorophenol monooxygenase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, P-NITROPHENOL | | Authors: | Pimviriyakul, P, Jaruwat, A, Chitnumsub, P, Chaiyen, P. | | Deposit date: | 2021-03-02 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into a flavin-dependent dehalogenase HadA explain catalysis and substrate inhibition via quadruple pi-stacking.
J.Biol.Chem., 297, 2021
|
|
2BSG
 
 | | The modeled structure of fibritin (gpwac) of bacteriophage T4 based on cryo-EM reconstruction of the extended tail of bacteriophage T4 | | Descriptor: | FIBRITIN | | Authors: | Kostyuchenko, V.A, Chipman, P.R, Leiman, P.G, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2005-05-20 | | Release date: | 2005-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | The Tail Structure of Bacteriophage T4 and its Mechanism of Contraction.
Nat.Struct.Mol.Biol., 12, 2005
|
|
7ETC
 
 | | Crystal structure of AbHpaI-Zn-(4R)-KDGal complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | (4R,5R)-4,5,6-tris(oxidanyl)-2-oxidanylidene-hexanoic acid, 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETF
 
 | | Crystal structure of AbHpaI-Mn-pyruvate-succinic semialdehyde complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, 4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETB
 
 | | Crystal structure of AbHpaI-Mn-pyruvate complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ET9
 
 | | Crystal structure of AbHpaI-Zn-pyruvate complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, PYRUVIC ACID, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ET8
 
 | | Crystal structure of thermostable AbHpaI, a class II metal dependent pyruvate aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETG
 
 | | Crystal structure of AbHpaI-Co-pyruvate-succinic semialdehyde complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, 4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETH
 
 | | Crystal structure of AbHpaI-Zn-pyruvate-propionaldehyde complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, PROPANAL, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETD
 
 | | Crystal structure of AbHpaI-Zn-(4S)-KDGlu complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
7ETE
 
 | | Crystal structure of AbHpaI-Mg-(4R)-KDGal complex, Class II aldolase, HpaI from Acinetobacter baumannii | | Descriptor: | (4R,5R)-4,5,6-tris(oxidanyl)-2-oxidanylidene-hexanoic acid, 4-hydroxy-2-oxoheptanedioate aldolase, CALCIUM ION, ... | | Authors: | Watthaisong, P, Binlaeh, A, Jaruwat, A, Chaiyen, P, Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic and structural insights into a stereospecific and thermostable Class II aldolase HpaI from Acinetobacter baumannii.
J.Biol.Chem., 297, 2021
|
|
