6NLQ
 
 | |
6NOW
 
 | |
8VO2
 
 | | Pathogenesis related 10-10 C59S mutant | | Descriptor: | Pathogenesis related 10-10 C59S mutant | | Authors: | Carr, S.C, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of a ligand-triggered intermolecular disulfide switch in a major latex protein from opium poppy.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7R7S
 
 | | p47-bound p97-R155H mutant with ATPgammaS | | Descriptor: | NSFL1 cofactor p47, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7R7T
 
 | | p47-bound p97-R155H mutant with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NSFL1 cofactor p47, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7R7U
 
 | | D1 and D2 domain structure of the p97(R155H)-p47 complex | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
8CNZ
 
 | | mmLarE-[4Fe-4S] phased by Fe-SAD | | Descriptor: | CHLORIDE ION, IODIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Pecqueur, L, Zecchin, P, Golinelli-Pimpaneau, B. | | Deposit date: | 2023-02-24 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-based insights into the mechanism of [4Fe-4S]-dependent sulfur insertase LarE.
Protein Sci., 33, 2024
|
|
6P3O
 
 | | Tetrahydroprotoberberine N-methyltransferase in complex with (S)-cis-N-methylstylopine and S-adenosylhomocysteine | | Descriptor: | (5S,12bS)-5-methyl-6,7,12b,13-tetrahydro-2H,4H,10H-[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquinolino[3,2-a]isoquinolin-5-ium, S-ADENOSYL-L-HOMOCYSTEINE, Tetrahydroprotoberberine N-methyltransferase | | Authors: | Lang, D.E, Morris, J.S, Rowley, M, Torres, M.A, Maksimovich, V.A, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2019-05-24 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function studies of tetrahydroprotoberberineN-methyltransferase reveal the molecular basis of stereoselective substrate recognition.
J.Biol.Chem., 294, 2019
|
|
8VO1
 
 | | Pathogenesis related 10-10 C155S mutant | | Descriptor: | Pathogenesis related 10-10 C155S mutant | | Authors: | Carr, S.C, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a ligand-triggered intermolecular disulfide switch in a major latex protein from opium poppy.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4D0W
 
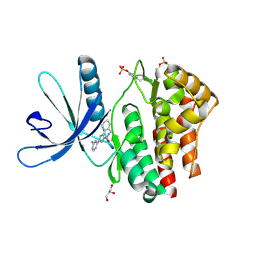 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 5-(2-aminopyrimidin-4-yl)-2-(5-chloro-2-methylphenyl)-1H-pyrrole-3-carboxamide, GLYCEROL, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Bertrand, J, Canevari, G, Fasolini, M, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
4D1S
 
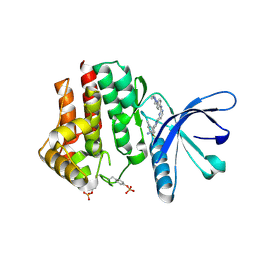 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 2-(5-chloro-2-methylphenyl)-1-methyl-5-(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1H-pyrrole-3-carboxamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Bertrand, J, Canevari, G, Fasolini, M, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-05-05 | | Release date: | 2014-07-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
9BOQ
 
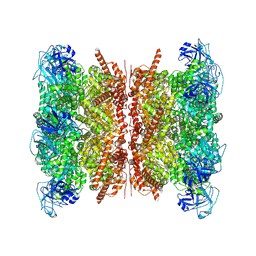 | | Human p97/VCP structure with a triazole inhibitor (NSC799462/dodecamer) | | Descriptor: | 3-(2,6-difluoro-4-{[(4P)-5-{[(2S)-hexan-2-yl]sulfanyl}-4-(pyridin-3-yl)-4H-1,2,4-triazol-3-yl]methoxy}phenyl)prop-2-yn-1-yl (1-methylpiperidin-4-yl)carbamate, ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, DeVore, K, Chiu, P.-L. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Mechanism of allosteric inhibition of human p97/VCP ATPase and its disease mutant by triazole inhibitors.
Commun Chem, 7, 2024
|
|
4D0X
 
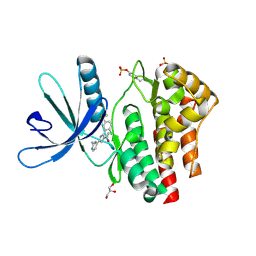 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 5-(2-aminopyrimidin-4-yl)-2-[2-chloro-5-(trifluoromethyl)phenyl]-1H-pyrrole-3-carboxamide, GLYCEROL, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Canevari, G, Fasolini, M, Bertrand, J, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
5KOC
 
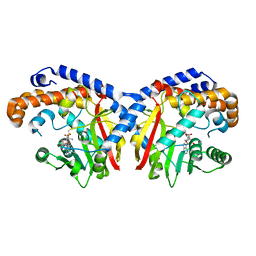 | | Pavine N-methyltransferase in complex with S-adenosylmethionine pH 7 | | Descriptor: | Pavine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Torres, M.A, Hoffarth, E, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
5KOK
 
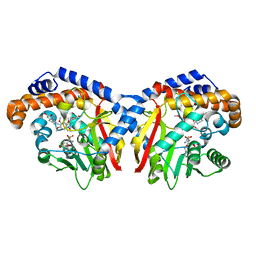 | | Pavine N-methyltransferase in complex with Tetrahydropapaverine and S-adenosylhomocysteine pH 7.25 | | Descriptor: | (1~{R})-1-[(3,4-dimethoxyphenyl)methyl]-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline, (1~{S})-1-[(3,4-dimethoxyphenyl)methyl]-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline, Pavine N-methyltransferase, ... | | Authors: | Torres, M.A, Hoffarth, E, Eugenio, L, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
5KPC
 
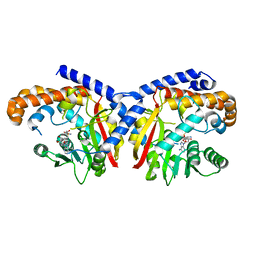 | | Pavine N-methyltransferase H206A mutant in complex with S-adenosylmethionine pH 6 | | Descriptor: | Pavine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Torres, M.A, Hoffarth, E, Eugenio, L, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-07-03 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
5KN4
 
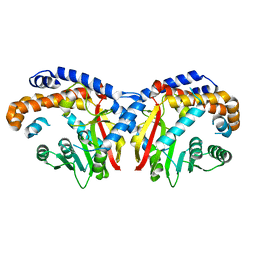 | | Pavine N-methyltransferase apoenzyme pH 6.0 | | Descriptor: | Pavine N-methyltransferase | | Authors: | Torres, M.A, Hoffarth, E, Eugenio, L, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-06-27 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
5KPG
 
 | | Pavine N-methyltransferase in complex with S-adenosylhomocysteine pH 7 | | Descriptor: | Pavine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Torres, M.A, Hoffarth, E, Eugenio, L, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-07-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
5LBM
 
 | | The asymmetric tetrameric structure of the formaldehyde sensing transcriptional repressor FrmR from Escherichia coli | | Descriptor: | FORMYL GROUP, Transcriptional repressor FrmR | | Authors: | Bisson, C, Baker, P.J, Green, J, Chivers, P.T. | | Deposit date: | 2016-06-16 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The mechanism of a formaldehyde-sensing transcriptional regulator.
Sci Rep, 6, 2016
|
|
1EIS
 
 | | UDA UNCOMPLEXED FORM. CRYSTAL STRUCTURE OF URTICA DIOICA AGGLUTININ, A SUPERANTIGEN PRESENTED BY MHC MOLECULES OF CLASS I AND CLASS II | | Descriptor: | PROTEIN (AGGLUTININ ISOLECTIN VI/AGGLUTININ ISOLECTIN V) | | Authors: | Saul, F.A, Rovira, P, Boulot, G, Van Damme, E.J.M, Peumans, W.J, Truffa-Bachi, P, Bentley, G.A. | | Deposit date: | 2000-02-28 | | Release date: | 2000-06-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II.
Structure Fold.Des., 8, 2000
|
|
1EN2
 
 | | UDA TETRASACCHARIDE COMPLEX. CRYSTAL STRUCTURE OF URTICA DIOICA AGGLUTININ, A SUPERANTIGEN PRESENTED BY MHC MOLECULES OF CLASS I AND CLASS II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ ISOLECTIN I/AGGLUTININ ISOLECTIN V/ AGGLUTININ ISOLECTIN VI | | Authors: | Saul, F.A, Rovira, P, Boulot, G, Van Damme, E.J.M, Peumans, W.J, Truffa-Bachi, P, Bentley, G.A. | | Deposit date: | 2000-03-20 | | Release date: | 2000-06-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II.
Structure Fold.Des., 8, 2000
|
|
1ENM
 
 | | UDA TRISACCHARIDE COMPLEX. CRYSTAL STRUCTURE OF URTICA DIOICA AGGLUTININ, A SUPERANTIGEN PRESENTED BY MHC MOLECULES OF CLASS I AND CLASS II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ ISOLECTIN I/AGGLUTININ ISOLECTIN V/ AGGLUTININ ISOLECTIN VI | | Authors: | Saul, F.A, Rovira, P, Boulot, G, Van Damme, E.J.M, Peumans, W.J, Truffa-Bachi, P, Bentley, G.A. | | Deposit date: | 2000-03-21 | | Release date: | 2000-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II.
Structure Fold.Des., 8, 2000
|
|
7RWL
 
 | | Envelope-associated Adeno-associated virus serotype 2 | | Descriptor: | Capsid protein VP1 | | Authors: | Hull, J.A, Mietzsch, M, Chipman, P, Strugatsky, D, McKenna, R. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural characterization of an envelope-associated adeno-associated virus type 2 capsid.
Virology, 565, 2021
|
|
6JHM
 
 | |
7RWT
 
 | | Adeno-associated virus type 2 | | Descriptor: | Capsid protein VP1 | | Authors: | Hull, J.A, Mietzsch, M, Chipman, P, Strugatsky, D, McKenna, R. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Structural characterization of an envelope-associated adeno-associated virus type 2 capsid.
Virology, 565, 2021
|
|
