3VH6
 
 | | Crystal structure of the chicken CENP-T histone fold/CENP-W/CENP-S/CENP-X heterotetrameric complex, crystal form II | | 分子名称: | CENP-S, CENP-T, CENP-W, ... | | 著者 | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | 登録日 | 2011-08-23 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.351 Å) | | 主引用文献 | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold
Cell(Cambridge,Mass.), 148, 2012
|
|
3WXQ
 
 | | Serial femtosecond X-ray structure of human fatty acid-binding protein type-3 (FABP3) in complex with stearic acid (C18:0) determined using X-ray free-electron laser at SACLA | | 分子名称: | Fatty acid-binding protein, heart, STEARIC ACID | | 著者 | Mizohata, E, Suzuki, M, Kakinouchi, K, Sugiyama, S, Murata, M, Sugahara, M, Nango, E, Tanaka, T, Tanaka, R, Tono, K, Song, C, Hatsui, T, Joti, Y, Yabashi, M, Iwata, S. | | 登録日 | 2014-08-04 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
6JSN
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(5R)-3-amino-5-methyl-9,9-dioxo-2,9lambda6-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | 分子名称: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | 著者 | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | 登録日 | 2019-04-08 | | 公開日 | 2019-08-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
4GPU
 
 | |
1EQ4
 
 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-04-03 | | 公開日 | 2000-04-19 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
6JSE
 
 | | Crystal Structure of BACE1 in complex with N-(3-((4S,5R)-2-amino-4-methyl-5-phenyl-5,6-dihydro-4H-1,3-thiazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | 分子名称: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | 著者 | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | 登録日 | 2019-04-08 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
1GTL
 
 | | The thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Pro-Phe-cho | | 分子名称: | ALDEHYDE INHIBITOR, CALCIUM ION, KUMAMOLYSIN, ... | | 著者 | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | 登録日 | 2002-01-16 | | 公開日 | 2002-06-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GT9
 
 | | High resolution crystal structure of a thermostable serine-carboxyl type proteinase, kumamolisin (kscp) | | 分子名称: | CALCIUM ION, KUMAMOLYSIN, SULFATE ION | | 著者 | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | 登録日 | 2002-01-14 | | 公開日 | 2002-06-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1SMY
 
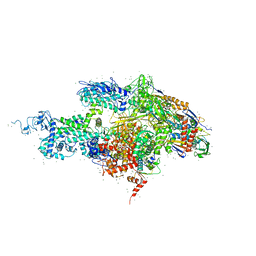 | | Structural basis for transcription regulation by alarmone ppGpp | | 分子名称: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | 著者 | Artsimovitch, I, Patlan, V, Sekine, S, Vassylyeva, M.N, Hosaka, T, Ochi, K, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-03-10 | | 公開日 | 2004-05-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis for transcription regulation by alarmone ppGpp
Cell(Cambridge,Mass.), 117, 2004
|
|
3X26
 
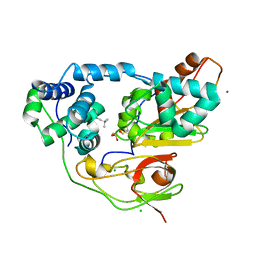 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 5 min | | 分子名称: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | 著者 | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | 登録日 | 2014-12-10 | | 公開日 | 2016-01-27 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1GTJ
 
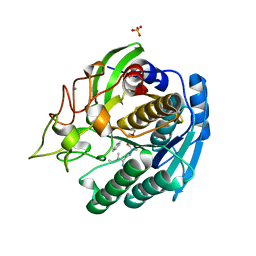 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Ala-Phe-cho | | 分子名称: | ALDEHYDE INHIBITOR, CALCIUM ION, KUMAMOLYSIN, ... | | 著者 | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | 登録日 | 2002-01-15 | | 公開日 | 2002-06-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
3X25
 
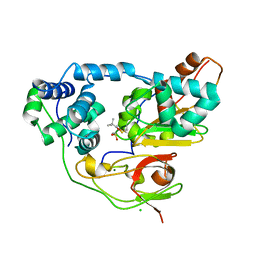 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 700 min | | 分子名称: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | 著者 | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | 登録日 | 2014-12-10 | | 公開日 | 2016-01-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1GTG
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolysin (kscp) | | 分子名称: | CALCIUM ION, KUMAMOLYSIN | | 著者 | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | 登録日 | 2002-01-15 | | 公開日 | 2002-06-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
3X24
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 120 min | | 分子名称: | 2,2-dimethylpropanenitrile, FE (III) ION, MAGNESIUM ION, ... | | 著者 | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | 登録日 | 2014-12-10 | | 公開日 | 2016-01-27 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1F54
 
 | | SOLUTION STRUCTURE OF THE APO N-TERMINAL DOMAIN OF YEAST CALMODULIN | | 分子名称: | CALMODULIN | | 著者 | Ishida, H, Takahashi, K, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | 登録日 | 2000-06-13 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structures of the N-terminal Domain of Yeast Calmodulin:
Ca2+-Dependent Conformational Change and Its Functional Implication
Biochemistry, 39, 2000
|
|
1F55
 
 | | SOLUTION STRUCTURE OF THE CALCIUM BOUND N-TERMINAL DOMAIN OF YEAST CALMODULIN | | 分子名称: | CALCIUM ION, CALMODULIN | | 著者 | Ishida, H, Takahashi, K, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | 登録日 | 2000-06-13 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structures of the N-terminal Domain of Yeast Calmodulin:
Ca2+-Dependent Conformational Change and Its Functional Implication
Biochemistry, 39, 2000
|
|
1IQW
 
 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF THE MOUSE ANTI-HUMAN FAS ANTIBODY HFE7A | | 分子名称: | ANTIBODY M-HFE7A, HEAVY CHAIN, LIGHT CHAIN | | 著者 | Ito, S, Takayama, T, Hanzawa, H, Ichikawa, K, Ohsumi, J, Serizawa, N, Hata, T, Haruyama, H. | | 登録日 | 2001-08-10 | | 公開日 | 2002-01-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the antigen-binding fragment of apoptosis-inducing mouse anti-human Fas monoclonal antibody HFE7A.
J.Biochem., 131, 2002
|
|
1IX5
 
 | | Solution structure of the Methanococcus thermolithotrophicus FKBP | | 分子名称: | FKBP | | 著者 | Suzuki, R, Nagata, K, Kawakami, M, Nemoto, N, Furutani, M, Adachi, K, Maruyama, T, Tanokura, M. | | 登録日 | 2002-06-12 | | 公開日 | 2003-06-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional Solution Structure of an Archaeal FKBP with a Dual Function of Peptidyl Prolyl cis-trans Isomerase and Chaperone-like Activities
J.MOL.BIOL., 328, 2003
|
|
1X1K
 
 | | Host-guest peptide (Pro-Pro-Gly)4-(Pro-alloHyp-Gly)-(Pro-Pro-Gly)4 | | 分子名称: | Host-guest peptide (Pro-Pro-Gly)4-(Pro-alloHyp-Gly)-(Pro-Pro-Gly)4 | | 著者 | Jiravanichanun, N, Hongo, C, Wu, G, Noguchi, K, Okuyama, K, Nishino, N, Silva, T. | | 登録日 | 2005-04-05 | | 公開日 | 2005-06-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Unexpected puckering of hydroxyproline in the guest triplets, hyp-pro-gly and pro-allohyp-gly sandwiched between pro-pro-gly sequence
Chembiochem, 6, 2005
|
|
1QWF
 
 | | C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND VSL12 | | 分子名称: | TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC, VAL-SER-LEU-ALA-ARG-ARG-PRO-LEU-PRO-PRO-LEU-PRO | | 著者 | Feng, S, Chiyoshi, K, Rickles, R.J, Schreiber, S.L. | | 登録日 | 1995-11-09 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1QWE
 
 | | C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND APP12 | | 分子名称: | ALA-PRO-PRO-LEU-PRO-PRO-ARG-ASN-ARG-PRO-ARG-LEU, TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC | | 著者 | Feng, S, Chiyoshi, K, Rickles, R.J, Schreiber, S.L. | | 登録日 | 1995-11-09 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1CXZ
 
 | | CRYSTAL STRUCTURE OF HUMAN RHOA COMPLEXED WITH THE EFFECTOR DOMAIN OF THE PROTEIN KINASE PKN/PRK1 | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, PROTEIN (HIS-TAGGED TRANSFORMING PROTEIN RHOA(0-181)), ... | | 著者 | Maesaki, R, Ihara, K, Shimizu, T, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | 登録日 | 1999-08-31 | | 公開日 | 1999-10-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The structural basis of Rho effector recognition revealed by the crystal structure of human RhoA complexed with the effector domain of PKN/PRK1.
Mol.Cell, 4, 1999
|
|
3WN8
 
 | | Crystal Structure of Collagen-Model Peptide, (POG)3-PRG-(POG)4 | | 分子名称: | collagen-like peptide | | 著者 | Okuyama, K, Haga, M, Noguchi, K, Tanaka, T. | | 登録日 | 2013-12-06 | | 公開日 | 2014-08-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Preferred side-chain conformation of arginine residues in a triple-helical structure.
Biopolymers, 101, 2014
|
|
1FGG
 
 | | CRYSTAL STRUCTURE OF 1,3-GLUCURONYLTRANSFERASE I (GLCAT-I) COMPLEXED WITH GAL-GAL-XYL, UDP, AND MN2+ | | 分子名称: | GLUCURONYLTRANSFERASE I, MANGANESE (II) ION, UNKNOWN ATOM OR ION, ... | | 著者 | Pedersen, L.C, Tsuchida, K, Kitagawa, H, Sugahara, K, Darden, T.A. | | 登録日 | 2000-07-28 | | 公開日 | 2001-01-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Heparan/chondroitin sulfate biosynthesis. Structure and mechanism of human glucuronyltransferase I.
J.Biol.Chem., 275, 2000
|
|
1IE6
 
 | | SOLUTION STRUCTURE OF IMPERATOXIN A | | 分子名称: | IMPERATOXIN A | | 著者 | Lee, C.W, Takeuchi, K, Takahashi, H, Sato, K, Shimada, I, Kim, D.H, Kim, J.I. | | 登録日 | 2001-04-07 | | 公開日 | 2003-06-10 | | 最終更新日 | 2024-10-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular basis of the high-affinity activation of type 1 ryanodine receptors by imperatoxin A.
Biochem.J., 377, 2004
|
|
