1IDV
 
 | |
1EF2
 
 | | CRYSTAL STRUCTURE OF MANGANESE-SUBSTITUTED KLEBSIELLA AEROGENES UREASE | | Descriptor: | MANGANESE (II) ION, UREASE ALPHA SUBUNIT, UREASE BETA SUBUNIT, ... | | Authors: | Yamaguchi, K, Cosper, N.J, Stalhandske, C, Scott, R.A, Pearson, M.A, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 2000-02-05 | | Release date: | 2000-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of metal-substituted Klebsiella aerogenes urease.
J.Biol.Inorg.Chem., 4, 1999
|
|
1R4H
 
 | | NMR Solution structure of the IIIc domain of GB Virus B IRES Element | | Descriptor: | 5'-R(*GP*GP*GP*CP*AP*AP*GP*CP*CP*C)-3' | | Authors: | Kaluarachchi, K, Thiviyanathan, V, Rijinbrand, R, Lemon, S.M, Gorenstein, D.G. | | Deposit date: | 2003-10-06 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mutational and structural analysis of stem-loop IIIC of the hepatitis C virus and GB virus B internal ribosome entry sites.
J.Mol.Biol., 343, 2004
|
|
1K9L
 
 | |
3WIT
 
 | |
1K9H
 
 | | NMR structure of DNA TGTGAGCGCTCACA | | Descriptor: | 5'-D(*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*A)-3' | | Authors: | Kaluarachchi, K, Gorenstein, D.G, Luxon, B.A. | | Deposit date: | 2001-10-29 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | How Do Proteins Recognize DNA? Solution Structure and Local Conformational Dynamics of Lac Operators by 2D NMR
J.Biomol.Struct.Dyn., Conversation 11, 2000
|
|
2E11
 
 | | The Crystal Structure of XC1258 from Xanthomonas campestris: A CN-hydrolase Superfamily Protein with an Arsenic Adduct in the Active Site | | Descriptor: | CACODYLATE ION, Hydrolase | | Authors: | Chin, K.-H, Tsai, Y.-D, Chan, N.-L, Huang, K.-F, Wang, A.H.-J, Chou, S.-H. | | Deposit date: | 2006-10-17 | | Release date: | 2007-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The crystal structure of XC1258 from Xanthomonas campestris: A putative procaryotic Nit protein with an arsenic adduct in the active site
Proteins, 69, 2007
|
|
3ATY
 
 | | Crystal structure of TcOYE | | Descriptor: | FLAVIN MONONUCLEOTIDE, Prostaglandin F2a synthase | | Authors: | Yamaguchi, K, Okamoto, N, Tokuoka, K, Sugiyama, S, Uchiyama, N, Matsumura, H, Inaka, K, Urade, Y, Inoue, T. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the stereoselective production of PGF2(alpha) by Old Yellow Enzyme from Trypanosoma cruzi
J.Biochem., 150, 2011
|
|
2DDX
 
 | |
3RZ4
 
 | |
2E12
 
 | | The crystal structure of XC5848 from Xanthomonas campestris adopting a novel variant of Sm-like motif | | Descriptor: | Hypothetical protein XCC3642 | | Authors: | Chin, K.-H, Ruan, S.-K, Wang, A.H.-J, Chou, S.-H. | | Deposit date: | 2006-10-17 | | Release date: | 2007-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | XC5848, an ORFan protein from Xanthomonas campestris, adopts a novel variant of Sm-like motif
Proteins, 68, 2007
|
|
4AUR
 
 | |
4AE3
 
 | | Crystal structure of ammosamide 272:myosin-2 motor domain complex | | Descriptor: | 1,2-ETHANEDIOL, ADP ORTHOVANADATE, AMMOSAMIDE 272, ... | | Authors: | Chinthalapudi, K, Heissler, S.M, Fenical, W, Manstein, D.J. | | Deposit date: | 2012-01-05 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Ammosamide Mediated Myosin Motor Activity Inhibition
To be Published
|
|
2FA5
 
 | | The crystal structure of an unliganded multiple antibiotic-resistance repressor (MarR) from Xanthomonas campestris | | Descriptor: | CHLORIDE ION, transcriptional regulator marR/emrR family | | Authors: | Chin, K.H, Tu, Z.L, Li, J.N, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2005-12-06 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of XC1739: a putative multiple antibiotic-resistance repressor (MarR) from Xanthomonas campestris at 1.8 A resolution
Proteins, 65, 2006
|
|
2XEL
 
 | | Molecular Mechanism of Pentachloropseudilin Mediated Inhibition of Myosin Motor Activity | | Descriptor: | 2,4-DICHLORO-6-(3,4,5-TRICHLORO-1H-PYRROL-2YL)PHENOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Chinthalapudi, K, Taft, M.H, Martin, R, Hartmann, F.K, Heissler, S.M, Tsiavaliaris, G, Gutzeit, H.O, Knoelker, H.J, Coluccio, L.M, Fedorov, R, Manstein, D.J. | | Deposit date: | 2010-05-16 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism and Specificity of Pentachloropseudilin-Mediated Inhibition of Myosin Motor Activity.
J.Biol.Chem., 286, 2011
|
|
7K66
 
 | | Structure of Blood Coagulation Factor VIII in Complex with an Anti-C1 Domain Pathogenic Antibody Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A9 heavy chain, ... | | Authors: | Childers, K.C, Gish, J, Jarvis, L, Peters, S, Garrels, C, Smith, I.W, Spencer, H.T, Spiegel, P.C. | | Deposit date: | 2020-09-18 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | Structure of blood coagulation factor VIII in complex with an anti-C1 domain pathogenic antibody inhibitor.
Blood, 137, 2021
|
|
2XJ9
 
 | | Dimer Structure of the bacterial cell division regulator MipZ | | Descriptor: | MAGNESIUM ION, MIPZ, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Michie, K.A, Lowe, J. | | Deposit date: | 2010-07-02 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Localized Dimerization and Nucleoid Binding Drive Gradient Formation by the Bacterial Cell Division Inhibitor Mipz.
Mol.Cell, 46, 2012
|
|
2XJ4
 
 | |
2P0Y
 
 | | Crystal structure of Q88YI3_LACPL from Lactobacillus plantarum. Northeast Structural Genomics Consortium target LpR6 | | Descriptor: | Hypothetical protein lp_0780 | | Authors: | Benach, J, Chen, Y, Seetharaman, J, Chi, K.H, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Q88YI3_LACPL from Lactobacillus plantarum.
To be Published
|
|
4ZMU
 
 | | Dcsbis, a diguanylate cyclase from Pseudomonas aeruginosa | | Descriptor: | diguanylate cyclase | | Authors: | Chen, Y, Liu, C, Liu, S, Chi, K, Gu, L. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | crystal structure of Dcsbis from Pseudomonas aeruginosa
To Be Published
|
|
4ZMM
 
 | | GGDEF domain of Dcsbis complexed with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), diguanylate cyclase | | Authors: | Chen, Y, Liu, C, Liu, S, Chi, K, Gu, L. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal structure of Dcsbis GGDEF domain complexed with c-di-GMP
To Be Published
|
|
1LZ6
 
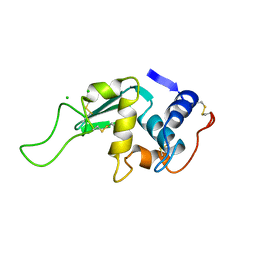 | | STRUCTURAL AND FUNCTIONAL ANALYSES OF THE ARG-GLY-ASP SEQUENCE INTRODUCED INTO HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, HUMAN LYSOZYME | | Authors: | Matsushima, M, Inaka, K, Yamada, T, Sekiguchi, K, Kikuchi, M. | | Deposit date: | 1993-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analyses of the Arg-Gly-Asp sequence introduced into human lysozyme.
J.Biol.Chem., 268, 1993
|
|
1VD8
 
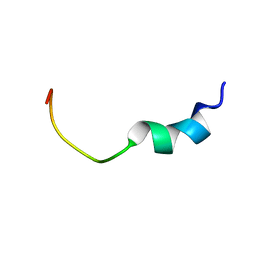 | | Solution structure of FMBP-1 tandem repeat 2 | | Descriptor: | fibroin-modulator-binding-protein-1 | | Authors: | Yamaki, T, Kawaguchi, K, Aizawa, T, Takiya, S, Demura, M, Nitta, K. | | Deposit date: | 2004-03-20 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FMBP-1 tandem repeat 2
To be Published
|
|
8IVG
 
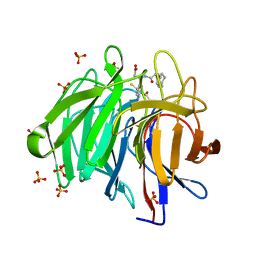 | | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1/Nrf2 PPI Inhibitor for Treatment of Chronic Kidney Disease | | Descriptor: | (3S)-3-(4-methylphenyl)-3-[2-(5,6,7,8-tetrahydronaphthalen-2-yl)ethanoylamino]propanoic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Kelch-like ECH-associated protein 1, ... | | Authors: | Otake, K, Ubukata, M, Nagahashi, N, Ogawa, N, Hantani, Y, Hantani, R, Adachi, T, Nomura, A, Yamaguchi, K, Maekawa, M, Mamada, H, Motomura, T, Sato, M, Harada, K. | | Deposit date: | 2023-03-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1-Nrf2 PPI Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
8IXS
 
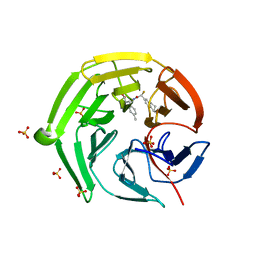 | | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1/Nrf2 PPI Inhibitor for Treatment of Chronic Kidney Disease | | Descriptor: | (2R,3S)-3-[[(2S)-2-fluoranyl-2-(5,6,7,8-tetrahydronaphthalen-2-yl)ethanoyl]amino]-2-methyl-3-(4-methylphenyl)propanoic acid, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Nomura, A, Yamaguchi, K, Adachi, T. | | Deposit date: | 2023-04-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1-Nrf2 PPI Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
