4COQ
 
 | | The complex of alpha-Carbonic anhydrase from Thermovibrio ammonificans with inhibitor sulfanilamide. | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CARBONATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | James, P, Isupov, M.N, Sayer, C, Berg, S, Lioliou, M, Kotlar, H, Littlechild, J.A. | | Deposit date: | 2014-01-30 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Structure of a Tetrameric [Alpha]-Carbonic Anhydrase from Thermovibrio Ammonificans Reveals a Core Formed Around Intermolecular Disulfides that Contribute to its Thermostability
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2N0Y
 
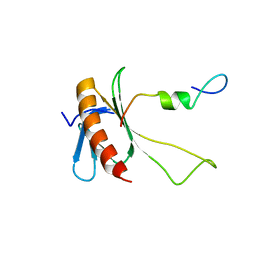 | | NMR structure of the complex between the C-terminal domain of the Rift Valley fever virus protein NSs and the PH domain of the Tfb1 subunit of TFIIH | | Descriptor: | Non-structural protein NS-S, RNA polymerase II transcription factor B subunit 1 | | Authors: | Cyr, N, de la Fuente, C, Lecoq, L, Guendel, I, Chabot, P.R, Kehn-Hall, K, Omichinski, J.G. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Omega XaV motif in the Rift Valley fever virus NSs protein is essential for degrading p62, forming nuclear filaments and virulence.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1HL7
 
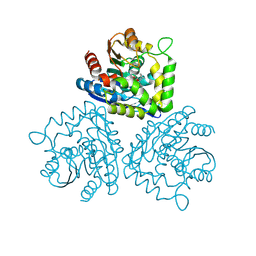 | | Gamma lactamase from an Aureobacterium species in complex with 3a,4,7,7a-tetrahydro-benzo [1,3] dioxol-2-one | | Descriptor: | 3A,4,7,7A-TETRAHYDRO-BENZO [1,3] DIOXOL-2-ONE, GAMMA LACTAMASE | | Authors: | Line, K, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2003-03-14 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of a (-)Gamma-Lactamase from an Aureobacterium Species Reveals a Tetrahedral Intermediate in the Active Site
J.Mol.Biol., 338, 2004
|
|
2N23
 
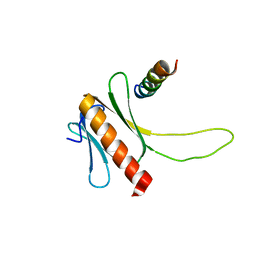 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the N-terminal activation domain of EKLF (TAD1) | | Descriptor: | Krueppel-like factor 1, RNA polymerase II transcription factor B subunit 1 | | Authors: | Lecoq, L, Morse, T, Raiola, L, Arseneault, G, Omichinski, J. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Complex between the N-terminal Transactivation Domain of EKLF and the p62/Tfb1 subunit of TFIIH
To be Published
|
|
1SAK
 
 | |
1SAE
 
 | |
1SAF
 
 | |
6ZZ2
 
 | | Cocktail experiment E: fragments 52, 58, and 63 at 90 mM concentration in complex with Endothiapepsin | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Hassaan, E, Klebe, G, Heine, A, Schiebel, J, Koester, H. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14885437 Å) | | Cite: | Cocktail experiment E: fragments 52, 58, and 63 at 90 mM concentration in complex with Endothiapepsin
To Be Published
|
|
7DOU
 
 | | Trimeric cement protein structure of Helicobacter pylori bacteriophage KHP40 | | Descriptor: | Cement protein gp16 | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
4C6H
 
 | | Haloalkane dehalogenase with 1-hexanol | | Descriptor: | CHLORIDE ION, HALOALKANE DEHALOGENASE, HEXAN-1-OL, ... | | Authors: | Novak, H.R, Sayer, C, Isupov, M, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Biochemical and Structural Characterisation of a Haloalkane Dehalogenase from a Marine Rhodobacteraceae.
FEBS Lett., 588, 2014
|
|
3ILE
 
 | | Crystal structure of ORF157-E86A of Acidianus filamentous virus 1 | | Descriptor: | NICKEL (II) ION, Putative uncharacterized protein | | Authors: | Goulet, A, Lichiere, J, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2009-08-07 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ORF157 from the archaeal virus Acidianus filamentous virus 1 defines a new class of nuclease
J.Virol., 84, 2010
|
|
1U0F
 
 | | Crystal structure of mouse phosphoglucose isomerase in complex with glucose 6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, BETA-MERCAPTOETHANOL, GLUCOSE-6-PHOSPHATE, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
1U0E
 
 | | Crystal structure of mouse phosphoglucose isomerase | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Glucose-6-phosphate isomerase, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
1U0G
 
 | | Crystal structure of mouse phosphoglucose isomerase in complex with erythrose 4-phosphate | | Descriptor: | BETA-MERCAPTOETHANOL, ERYTHOSE-4-PHOSPHATE, GLYCEROL, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
3K6W
 
 | | Apo and ligand bound structures of ModA from the archaeon Methanosarcina acetivorans | | Descriptor: | MOLYBDATE ION, SULFATE ION, Solute-binding protein MA_0280 | | Authors: | Chan, S, Chernishof, I, Giuroiu, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1S6L
 
 | | Solution structure of MerB, the Organomercurial Lyase involved in the bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase | | Authors: | Di Lello, P, Benison, G.C, Valafar, H, Pitts, K.E, Summers, A.O, Legault, P, Omichinski, J.G. | | Deposit date: | 2004-01-25 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies reveal a novel protein fold for MerB, the organomercurial lyase involved in the bacterial mercury resistance system.
Biochemistry, 43, 2004
|
|
8T31
 
 | |
2WDU
 
 | | Fasciola hepatica sigma class GST | | Descriptor: | BROMIDE ION, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Line, K, Isupov, M.N, LaCourse, E.J, Brophy, P.M, Littlechild, J.A. | | Deposit date: | 2009-03-26 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The 1.6 Angstrom Crystal Structure of the Fasciola Hepatica Sigma Class Gst
To be Published
|
|
3K6V
 
 | | M. acetivorans Molybdate-Binding Protein (ModA) in Citrate-Bound Open Form | | Descriptor: | CITRIC ACID, Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2WB9
 
 | | Fasciola hepatica sigma class GST | | Descriptor: | BROMIDE ION, CYSTEINE, GLUTATHIONE, ... | | Authors: | Line, K, Isupov, M.N, LaCourse, J, Brophy, P.M, Littlechild, J.A. | | Deposit date: | 2009-02-23 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The 1.6 Angstrom Crystal Structure of the Fasciola Hepatica Sigma Class Gst
To be Published
|
|
1SAL
 
 | |
3K6U
 
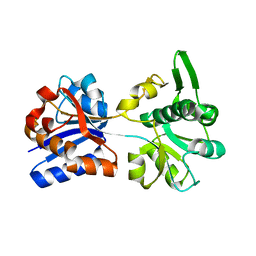 | | M. acetivorans Molybdate-Binding Protein (ModA) in Unliganded Open Form | | Descriptor: | Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4BRZ
 
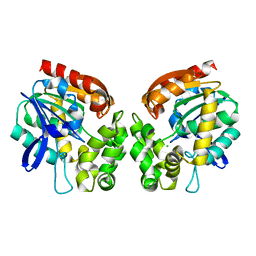 | | Haloalkane dehalogenase | | Descriptor: | CHLORIDE ION, HALOALKANE DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2013-06-06 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and Structural Characterisation of a Haloalkane Dehalogenase from a Marine Rhodobacteraceae.
FEBS Lett., 588, 2014
|
|
1UP8
 
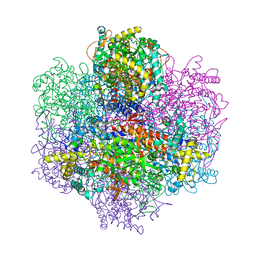 | | Recombinant vanadium-dependent bromoperoxidase from red algae Corallina pilulifera | | Descriptor: | CALCIUM ION, PHOSPHATE ION, VANADIUM-DEPENDENT BROMOPEROXIDASE 1 | | Authors: | Garcia-Rodriguez, E, Isupov, M, Ohshiro, T, Izumi, Y, Littlechild, J.A. | | Deposit date: | 2003-09-29 | | Release date: | 2003-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enhancing Effect of Calcium and Vanadium Ions on Thermal Stability of Bromoperoxidase from Corallina Pilulifera.
J.Biol.Inorg.Chem., 10, 2005
|
|
3HG0
 
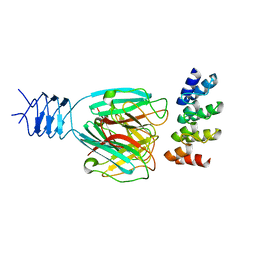 | | Crystal structure of a DARPin in complex with ORF49 from Lactococcal phage TP901-1 | | Descriptor: | Baseplate protein, Designed Ankyrin Repeat Protein (DARPin) 20 | | Authors: | Veesler, D, Dreier, B, Blangy, S, Lichiere, J, Tremblay, D, Moineau, S, Spinelli, S, Tegoni, M, Pluckthun, A, Campanacci, V, Cambillau, C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of a DARPin neutralizing inhibitor of lactococcal phage TP901-1: comparison of DARPin and camelid VHH binding mode.
J.Biol.Chem., 284, 2009
|
|
