7YUA
 
 | |
7YV0
 
 | |
7DMN
 
 | | Crystal structure of two pericyclases catalyzing [4+2] cycloaddition | | 分子名称: | Diels-Alderase fsa2, GLYCEROL | | 著者 | Chi, C.B, Wang, Z.D, Liu, T, Zhang, Z.Y, Ma, M. | | 登録日 | 2020-12-04 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of Fsa2 and Phm7 Catalyzing [4 + 2] Cycloaddition Reactions with Reverse Stereoselectivities in Equisetin and Phomasetin Biosynthesis.
Acs Omega, 6, 2021
|
|
7OSR
 
 | |
7OSW
 
 | |
8J5V
 
 | |
2MZU
 
 | |
2N0T
 
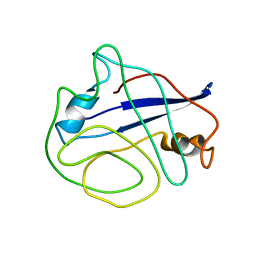 | | Structural ensemble of the enzyme cyclophilin reveals an orchestrated mode of action at atomic resolution | | 分子名称: | Peptidyl-prolyl cis-trans isomerase A | | 著者 | Chi, C.N, Voegeli, B, Bibow, S, Strotz, D, Orts, J, Guntert, P, Riek, R. | | 登録日 | 2015-03-13 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Structural Ensemble for the Enzyme Cyclophilin Reveals an Orchestrated Mode of Action at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5Y4U
 
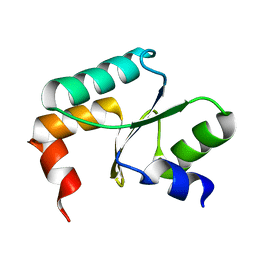 | | Crystal structure of Grx domain of Grx3 from Saccharomyces cerevisiae | | 分子名称: | Monothiol glutaredoxin-3 | | 著者 | Chi, C.B, Tang, Y.J, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | 登録日 | 2017-08-05 | | 公開日 | 2018-08-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J.Mol.Biol., 430, 2018
|
|
5Y4T
 
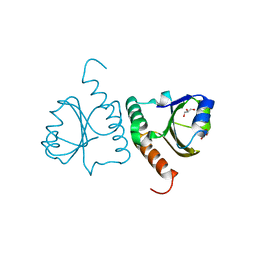 | | Crystal structure of Trx domain of Grx3 from Saccharomyces cerevisiae | | 分子名称: | GLYCEROL, Glutaredoxin | | 著者 | Chi, C.B, Tang, Y.J, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | 登録日 | 2017-08-05 | | 公開日 | 2018-08-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J.Mol.Biol., 430, 2018
|
|
3M5N
 
 | |
3M5O
 
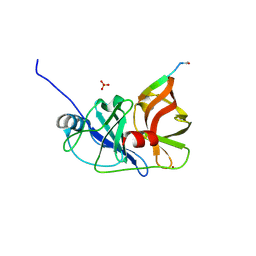 | |
3LZU
 
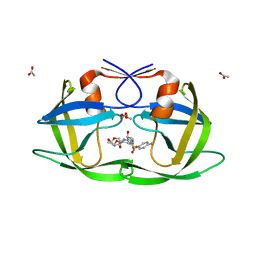 | | Crystal Structure of a Nelfinavir Resistant HIV-1 CRF01_AE Protease variant (N88S) in Complex with the Protease Inhibitor Darunavir. | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 protease | | 著者 | Schiffer, C.A, Bandaranayake, R.M. | | 登録日 | 2010-03-01 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
3LZV
 
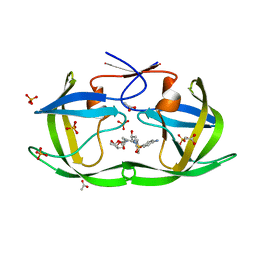 | | Structure of Nelfinavir-resistant HIV-1 protease (D30N/N88D) in complex with Darunavir. | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 Protease, ... | | 著者 | Schiffer, C.A, Kolli, M. | | 登録日 | 2010-03-01 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
7E22
 
 | |
4UMG
 
 | | Crystal structure of the Lin-41 filamin domain | | 分子名称: | PROTEIN LIN-41 | | 著者 | Tocchini, C, Keusch, J.J, Miller, S.B, Finger, S, Gut, H, Stadler, M, Ciosk, R. | | 登録日 | 2014-05-16 | | 公開日 | 2014-10-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | The Trim-Nhl Protein Lin-41 Controls the Onset of Developmental Plasticity in Caenorhabditis Elegans.
Plos Genet., 10, 2014
|
|
8T6Q
 
 | |
8SYG
 
 | |
1F7A
 
 | |
8T18
 
 | |
8T15
 
 | |
8T17
 
 | |
8T6K
 
 | |
4Y00
 
 | | Crystal Structure of Human TDP-43 RRM1 Domain with D169G Mutation in Complex with an Unmodified Single-stranded DNA | | 分子名称: | DNA (5'-D(P*TP*TP*GP*AP*GP*CP*GP*T)-3'), TAR DNA-binding protein 43 | | 著者 | Chiang, C.H, Kuo, P.H, Yang, W.Z, Yuan, H.S. | | 登録日 | 2015-02-05 | | 公開日 | 2016-02-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural analysis of disease-related TDP-43 D169G mutation: linking enhanced stability and caspase cleavage efficiency to protein accumulation
Sci Rep, 6, 2016
|
|
4Y0F
 
 | | Crystal Structure of Human TDP-43 RRM1 Domain in Complex with an Unmodified Single-stranded DNA | | 分子名称: | DNA (5'-D(*GP*TP*TP*GP*AP*GP*CP*GP*TP*T)-3'), TAR DNA-binding protein 43 | | 著者 | Chiang, C.H, Kuo, P.H, Doudeva, L.G, Wang, Y.T, Yuan, H.S. | | 登録日 | 2015-02-06 | | 公開日 | 2016-02-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.648 Å) | | 主引用文献 | Structural analysis of disease-related TDP-43 D169G mutation: linking enhanced stability and caspase cleavage efficiency to protein accumulation
Sci Rep, 6, 2016
|
|
