8TGF
 
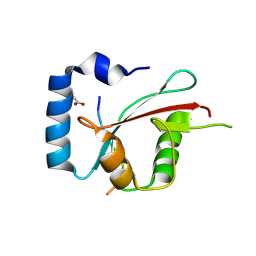 | | Crystal structure of cEPG5 LIR/LGG-1 complex | | Descriptor: | ACETATE ION, Ectopic P granules protein 5, Protein lgg-1 | | Authors: | Cheung, Y.W.S, Yip, C.K. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of the mode of interaction between the tandem LC3-interacting region motif of the autophagy tethering factor EPG5 and ATG8 proteins
To be published
|
|
7JHX
 
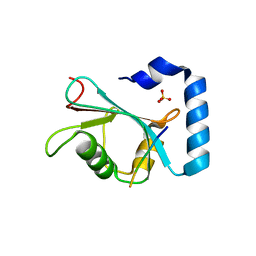 | | Crystal structure of hEPG5 LIR/GABARAPL1 complex | | Descriptor: | Ectopic P granules protein 5 homolog, Gamma-aminobutyric acid receptor-associated protein-like 1, SULFATE ION | | Authors: | Cheung, Y.W.S, Yip, C.K. | | Deposit date: | 2020-07-21 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Insights on autophagosome-lysosome tethering from structural and biochemical characterization of human autophagy factor EPG5.
Commun Biol, 4, 2021
|
|
1W2I
 
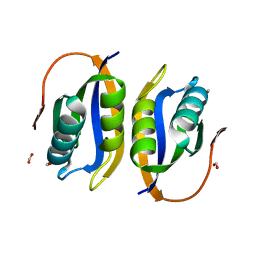 | | Crystal structuore of acylphosphatase from Pyrococcus horikoshii complexed with formate | | Descriptor: | ACYLPHOSPHATASE, FORMIC ACID | | Authors: | Cheung, Y.Y, Lam, S.Y, Chu, W.K, Allen, M.D, Bycroft, M, Wong, K.B. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Hyperthermophilic Archaeal Acylphosphatase from Pyrococcus Horikoshii-Structural Insights Into Enzymatic Catalysis, Thermostability, and Dimerization
Biochemistry, 44, 2005
|
|
3ZH2
 
 | | Structure of Plasmodium falciparum lactate dehydrogenase in complex with a DNA aptamer | | Descriptor: | DNA APTAMER, L-LACTATE DEHYDROGENASE | | Authors: | Cheung, Y.W, Kwok, J, Law, A.W.L, Watt, R.M, Kotaka, M, Tanner, J.A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Discriminatory Recognition of Plasmodium Lactate Dehydrogenase by a DNA Aptamer
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6TXR
 
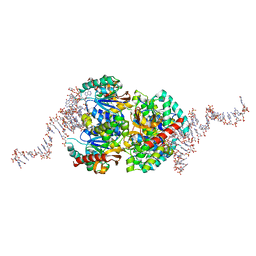 | | Structural insights into cubane-modified aptamer recognition of a malaria biomarker | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Cheung, Y, Roethlisberger, P, Mechaly, A, Weber, P, Wong, A, Lo, Y, Haouz, A, Savage, P, Hollenstein, M, Tanner, J. | | Deposit date: | 2020-01-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4FES
 
 | | Structure of OSH4 in complex with cholesterol analogs | | Descriptor: | (3S,8S,9S,10R,13S,14S,17R)-3-hydroxy-10,13-dimethyl-17-[(2S,6S)-6-methyl-3-oxooctan-2-yl]-1,2,3,4,7,8,9,10,11,12,13,14,15,17-tetradecahydro-16H-cyclopenta[a]phenanthren-16-one, Protein KES1 | | Authors: | Koag, M.C, Monzingo, A.F, Cheun, Y, Lee, S. | | Deposit date: | 2012-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structure of 16,22-diketocholesterol bound to oxysterol-binding protein Osh4.
Steroids, 78, 2013
|
|
4F4B
 
 | | Structure of OSH4 with a cholesterol analog | | Descriptor: | (3beta,9beta,25R)-3-hydroxy-26-iodocholest-5-ene-16,22-dione, Protein KES1 | | Authors: | Koag, M.C, Monzingo, A.F, Cheun, Y, Lee, S. | | Deposit date: | 2012-05-10 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Synthesis and structure of 16,22-diketocholesterol bound to oxysterol-binding protein Osh4.
Steroids, 78, 2013
|
|
