2JIY
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ALA M149 REPLACED WITH TRP (CHAIN M, AM149W) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Fyfe, P.K, Potter, J.A, Cheng, J, Williams, C.M, Watson, A.J, Jones, M.R. | | Deposit date: | 2007-07-03 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Responses to Cavity-Creating Mutations in an Integral Membrane Protein.
Biochemistry, 46, 2007
|
|
1L3M
 
 | | The Solution Structure of [d(CGC)r(amamam)d(TTTGCG)]2 | | Descriptor: | 5'-D(*CP*GP*C)-R(P*(A39)P*(A39)P*(A39))-D(P*TP*TP*TP*GP*CP*G)-3' | | Authors: | Tsao, Y.P, Wang, L.Y, Hsu, S.T, Jain, M.L, Chou, S.H, Huang, W.C, Cheng, J.W. | | Deposit date: | 2002-02-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of [d(CGC)r(amamam)d(TTTGCG)]2.
J.Biomol.NMR, 21, 2001
|
|
6TNU
 
 | | Yeast 80S ribosome in complex with eIF5A and decoding A-site and P-site tRNAs. | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
2JJ0
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ALA M248 REPLACED WITH TRP (CHAIN M, AM248W) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Fyfe, P.K, Potter, J.A, Cheng, J, Williams, C.M, Watson, A.J, Jones, M.R. | | Deposit date: | 2007-07-03 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Responses to Cavity-Creating Mutations in an Integral Membrane Protein.
Biochemistry, 46, 2007
|
|
5DEU
 
 | | Crystal structure of TET2-5hmC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DNA (5'-D(*AP*CP*CP*AP*CP*(5HC)P*GP*GP*TP*GP*GP*T)-3'), ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
5D9Y
 
 | | Crystal structure of TET2-5fC complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*(5FC)P*GP*AP*AP*GP*CP*T)-3'), DNA (5'-D(*AP*GP*CP*TP*TP*CP*GP*AP*CP*AP*GP*T)-3'), FE (III) ION, ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-19 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
175D
 
 | |
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
1BJH
 
 | | HAIRPIN LOOPS CONSISTING OF SINGLE ADENINE RESIDUES CLOSED BY SHEARED A(DOT)A AND G(DOT)G PAIRS FORMED BY THE DNA TRIPLETS AAA AND GAG: SOLUTION STRUCTURE OF THE D(GTACAAAGTAC) HAIRPIN, NMR, 16 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*AP*AP*AP*GP*TP*AP*C)-3') | | Authors: | Chou, S.-H, Zhu, L, Gao, Z, Cheng, J.-W, Reid, B.R. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Hairpin loops consisting of single adenine residues closed by sheared A.A and G.G pairs formed by the DNA triplets AAA and GAG: solution structure of the d(GTACAAAGTAC) hairpin.
J.Mol.Biol., 264, 1996
|
|
6LUQ
 
 | | Haloperidol bound D2 dopamine receptor structure inspired discovery of subtype selective ligands | | Descriptor: | 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one, OLEIC ACID, chimera of D(2) dopamine receptor and Endolysin | | Authors: | Fan, L, Tan, L, Chen, Z, Qi, J, Nie, F, Luo, Z, Cheng, J, Wang, S. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Haloperidol bound D2dopamine receptor structure inspired the discovery of subtype selective ligands.
Nat Commun, 11, 2020
|
|
4I8Y
 
 | | Structure of the unliganded N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ACETATE ION, CHLORIDE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-04 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
8JNG
 
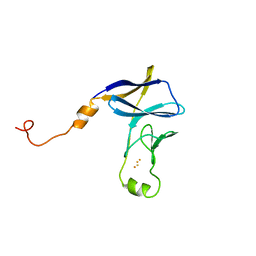 | | Methanesulfonate monooxygenase ferredoxin subunit of PBS-PSII-PSI-LHCs from Porphyridium purpureum. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Methanesulfonate monooxygenase ferredoxin subunit | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2023-06-06 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of lateral hexamer of PBS-PSII-PSI-LHCs megacomplex at 6.3 Angstroms resolution.
To Be Published
|
|
8JNN
 
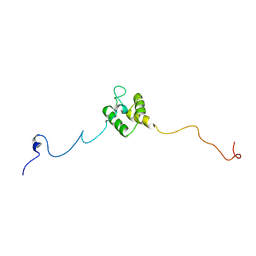 | | linker protein LPP1 from red algal Porphyridium purpureum. | | Descriptor: | LPP1 | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2023-06-06 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of lateral hexamer of PBS-PSII-PSI-LHCs megacomplex at 6.3 Angstroms resolution.
To Be Published
|
|
8JNM
 
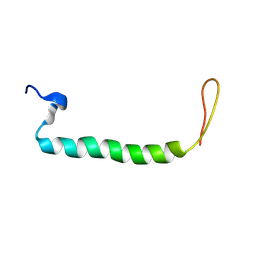 | | PsbW from red algal Porphyridium purpureum. | | Descriptor: | PsbW | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2023-06-06 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of lateral hexamer of PBS-PSII-PSI-LHCs megacomplex at 6.3 Angstroms resolution.
To Be Published
|
|
8JNL
 
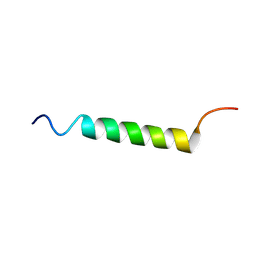 | | Psb34 from red algal Porphyridium purpureum. | | Descriptor: | Psb34 | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2023-06-06 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of lateral hexamer of PBS-PSII-PSI-LHCs megacomplex at 6.3 Angstroms resolution.
To Be Published
|
|
4I9J
 
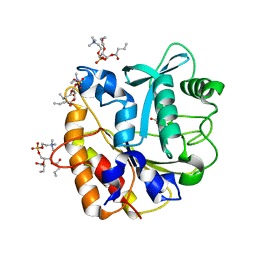 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus bound to diC4PC | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I90
 
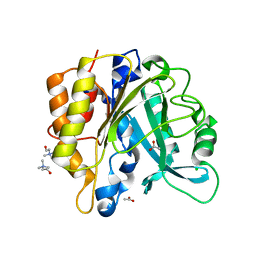 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus bound to choline | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-04 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I9M
 
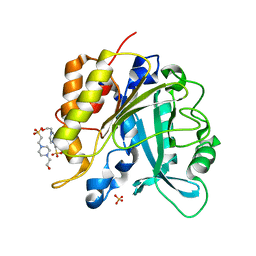 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from Staphylococcus aureus bound to HEPES | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
8I9P
 
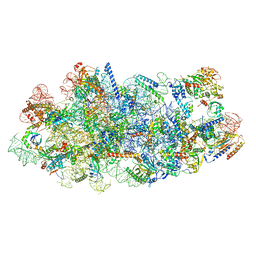 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Mak16 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, 60S ribosomal protein L16-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9X
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - Ytm1-1 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9Z
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Spb4 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9T
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Dbp10-1 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9R
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State 5S RNP | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, 60S ribosomal protein L16-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8IA0
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Puf6 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9V
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Dbp10-2 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
