8K0A
 
 | |
8K04
 
 | | CryoEM structure of a 2,3-hydroxycinnamic acid 1,2-dioxygenase MhpB in apo form | | Descriptor: | 2,3-dihydroxyphenylpropionate/2,3-dihydroxicinnamic acid 1,2-dioxygenase | | Authors: | Jiang, W.X, Cheng, X.Q, Ma, L.X, Xing, Q. | | Deposit date: | 2023-07-07 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | CryoEM structure of the NADP-dependent malic enzyme in complex with oxaloacetate
To Be Published
|
|
8JZO
 
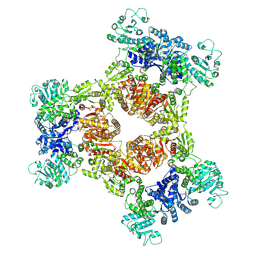 | |
6UKH
 
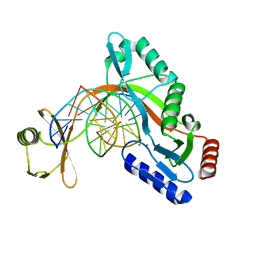 | | HhaI endonuclease in Complex with DNA in space group P41212 (pH 6.0) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*CP*AP*AP*GP*CP*GP*CP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*TP*GP*CP*GP*CP*TP*TP*GP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2019-10-04 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structure of HhaI endonuclease with cognate DNA at an atomic resolution of 1.0 angstrom.
Nucleic Acids Res., 48, 2020
|
|
6DQ4
 
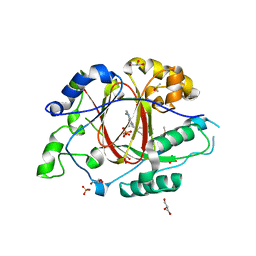 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR GSK-J1 | | Descriptor: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
7SFF
 
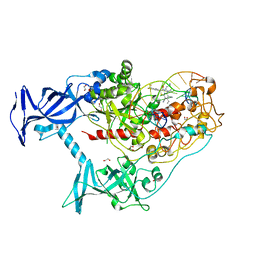 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3852279B | | Descriptor: | (2R)-2-{[6-(4-aminopiperidin-1-yl)-3,5-dicyano-4-ethylpyridin-2-yl]sulfanyl}-2-[4-(trifluoromethyl)phenyl]acetamide, 1,2-ETHANEDIOL, DNA (12-MER), ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
7SFD
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3543105A | | Descriptor: | (2S)-2-({3,5-dicyano-4-ethyl-6-[4-(2-hydroxyethyl)-1,4-diazepan-1-yl]pyridin-2-yl}sulfanyl)-2-phenylacetamide, 1,2-ETHANEDIOL, DNA (12-MER), ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
7SFC
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3735967A | | Descriptor: | 1,2-ETHANEDIOL, DNA (12-MER), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
7SFE
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3830334A | | Descriptor: | (2R)-2-{[6-(4-aminopiperidin-1-yl)-3,5-dicyano-4-ethylpyridin-2-yl]amino}-2-phenylacetamide, 1,2-ETHANEDIOL, DNA (12-MER), ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
7SFG
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Cofactor SAM | | Descriptor: | 1,2-ETHANEDIOL, DNA (cytosine-5)-methyltransferase 1, DNA Strand 1, ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
6DQ8
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR N49 i.e. 2-((2-chlorophenyl)(2-(1-methylpyrrolidin-2-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-[(R)-(2-chlorophenyl){2-[(2S)-1-methylpyrrolidin-2-yl]ethoxy}methyl]thieno[3,2-b]pyridine-7-carboxylic acid, 2-[(S)-(2-chlorophenyl){2-[(2S)-1-methylpyrrolidin-2-yl]ethoxy}methyl]thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
7C7U
 
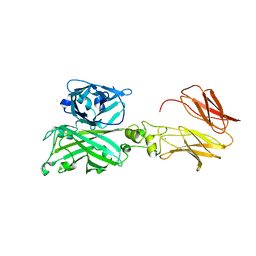 | | Biofilm associated protein - BSP domain | | Descriptor: | Biofilm-associated surface protein, CALCIUM ION | | Authors: | Ma, J.F, Xu, Z.H, Zhang, Y.K, Cheng, X, Fan, S.L, Wang, J.W, Fang, X.Y. | | Deposit date: | 2020-05-26 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural mechanism for modulation of functional amyloid and biofilm formation by Staphylococcal Bap protein switch.
Embo J., 40, 2021
|
|
7C7R
 
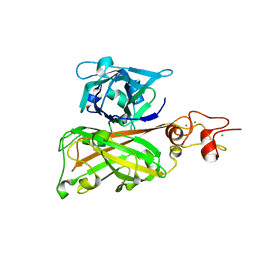 | | Biofilm associated protein - B domain | | Descriptor: | Biofilm-associated surface protein, CALCIUM ION | | Authors: | Ma, J.F, Xu, Z.H, Zhang, Y.K, Cheng, X, Fan, S.L, Wang, J.W, Fang, X.Y. | | Deposit date: | 2020-05-26 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural mechanism for modulation of functional amyloid and biofilm formation by Staphylococcal Bap protein switch.
Embo J., 40, 2021
|
|
6UKG
 
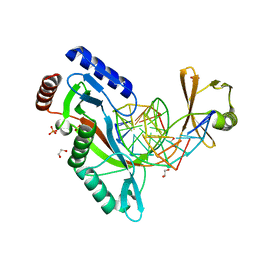 | | HhaI endonuclease in Complex With DNA in space group P21 (pH 4.2) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*AP*GP*CP*GP*CP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*TP*GP*CP*GP*CP*TP*TP*GP*GP*A)-3'), ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2019-10-04 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of HhaI endonuclease with cognate DNA at an atomic resolution of 1.0 angstrom.
Nucleic Acids Res., 48, 2020
|
|
6UKI
 
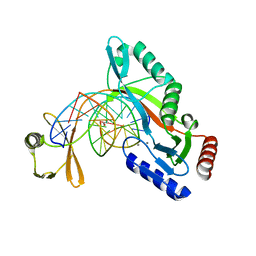 | |
6V62
 
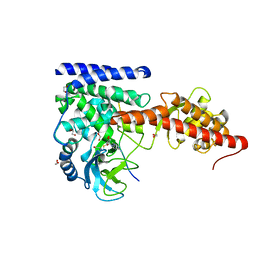 | | SETD3 double mutant (N255F/W273A) in Complex with an Actin Peptide with His73 Replaced with Lysine | | Descriptor: | 1,2-ETHANEDIOL, Actin, cytoplasmic 1, ... | | Authors: | Dai, S, Horton, J.R, Cheng, X. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | An engineered variant of SETD3 methyltransferase alters target specificity from histidine to lysine methylation.
J.Biol.Chem., 295, 2020
|
|
8JBZ
 
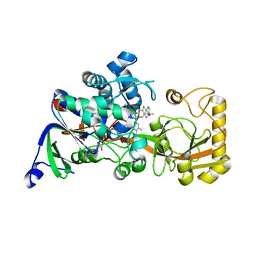 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 4-androstadiene-3,17- dione | | Descriptor: | 3-ketosteroid dehydrogenase, 4-ANDROSTENE-3-17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-05-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 4-androstadiene-3,17- dione
To Be Published
|
|
4EW4
 
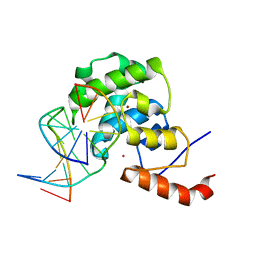 | | mouse MBD4 glycosylase domain in complex with DNA containing a ribose sugar | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*GP*(3DR)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4, ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.791 Å) | | Cite: | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
7CMV
 
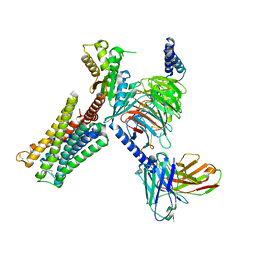 | | Dopamine Receptor D3R-Gi-PD128907 complex | | Descriptor: | (4aR,10bR)-4-propyl-3,4a,5,10b-tetrahydro-2H-chromeno[4,3-b][1,4]oxazin-9-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
7CMU
 
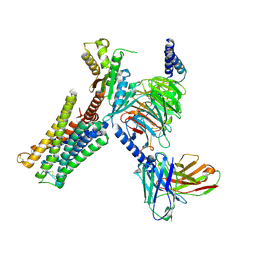 | | Dopamine Receptor D3R-Gi-Pramipexole complex | | Descriptor: | (6S)-N6-propyl-4,5,6,7-tetrahydro-1,3-benzothiazole-2,6-diamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
8KGZ
 
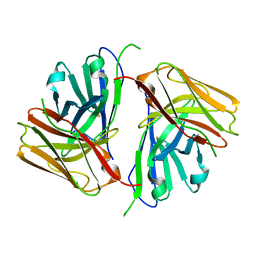 | | Crystal structure of single-chain Fv antibody against antigen peptide from SARS-CoV2 S-spike protein | | Descriptor: | Thioredoxin 1,scFv | | Authors: | Ma, Q.Q, Zhang, B.L, Cheng, X.Y, Huang, Y.Q, Su, Z.D. | | Deposit date: | 2023-08-20 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of sheep scFv antibody against an antigen peptide of SARS-CoV2 S-spike protein
To Be Published
|
|
5IVB
 
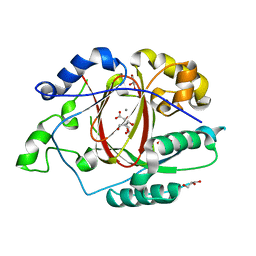 | |
5IVC
 
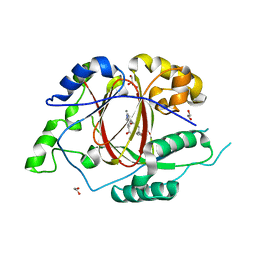 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N3 (4'-[(2-phenylethyl)carbamoyl][2,2'-bipyridine]-4-carboxylic acid) | | Descriptor: | 1,2-ETHANEDIOL, 4'-[(2-phenylethyl)carbamoyl][2,2'-bipyridine]-4-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IWF
 
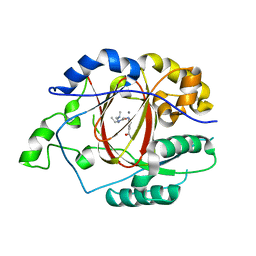 | |
5IVF
 
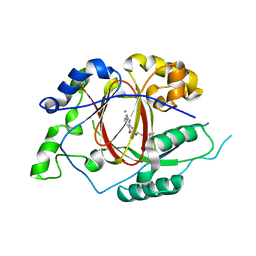 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N10 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol | | Descriptor: | 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol, Lysine-specific demethylase 5A, MANGANESE (II) ION | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
