3V6G
 
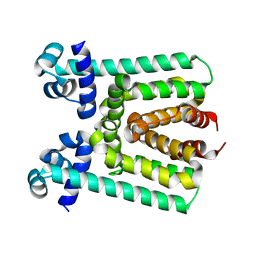 | | Crystal Structure of Transcriptional Regulator | | Descriptor: | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY DEOR-FAMILY) | | Authors: | Do, S.V, Bolla, J.R, Chen, X, Yu, E.W. | | Deposit date: | 2011-12-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis.
Nucleic Acids Res., 40, 2012
|
|
3V78
 
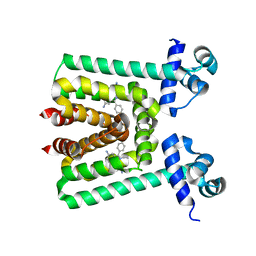 | | Crystal Structure of Transcriptional Regulator | | Descriptor: | ETHIDIUM, PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY DEOR-FAMILY) | | Authors: | Do, S.V, Bolla, J.R, Chen, X, Yu, E.W. | | Deposit date: | 2011-12-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis.
Nucleic Acids Res., 40, 2012
|
|
8UD4
 
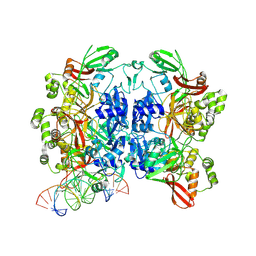 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, state 1 | | Descriptor: | Non-structural protein 15, RNA (35-MER) | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 15, 2024
|
|
8UD2
 
 | | SARS-CoV-2 Nsp15, apo-form | | Descriptor: | Non-structural protein 15 | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 15, 2024
|
|
8UD3
 
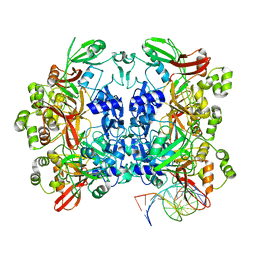 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, consensus form | | Descriptor: | Non-structural protein 15, RNA (35-MER) | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 15, 2024
|
|
3T6N
 
 | |
8W0A
 
 | |
8WCJ
 
 | | Crystal structure of GB3 penta mutation L5V/K10H/T16S/K19E/Y33I | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Qin, M.M, Chen, X.X, Zhang, X.Y, Song, X.F, Yao, L.S. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Protein Allostery Study in Cells Using NMR Spectroscopy.
Anal.Chem., 96, 2024
|
|
3HRO
 
 | | Crystal structure of a C-terminal coiled coil domain of Transient receptor potential (TRP) channel subfamily P member 2 (TRPP2, polycystic kidney disease 2) | | Descriptor: | Transient receptor potential (TRP) channel subfamily P member 2 (TRPP2), also called Polycystin-2 or polycystic kidney disease 2(PKD2) | | Authors: | Yu, Y, Ulbrich, M.H, Li, M.-H, Buraei, Z, Chen, X.-Z, Ong, A.C.M, Tong, L, Isacoff, E.Y, Yang, J. | | Deposit date: | 2009-06-09 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and molecular basis of the assembly of the TRPP2/PKD1 complex.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5IHZ
 
 | | Crystal structure of anti-gliadin 1002-1E01 Fab fragment | | Descriptor: | 1E01 Fab fragment heavy chain, 1E01 Fab fragment light chain | | Authors: | Snir, O, Chen, X, Gidoni, M, du Pre, M.F, Zhao, Y, Steinsbo, O, Lundin, K.E, Yaari, G, Sollid, L.M. | | Deposit date: | 2016-03-01 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Stereotyped antibody responses target posttranslationally modified gluten in celiac disease.
JCI Insight, 2, 2017
|
|
8UD5
 
 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, state 2 | | Descriptor: | Non-structural protein 15, RNA (35-MER) | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 15, 2024
|
|
5IK3
 
 | | Crystal structure of anti-gliadin 1002-1E03 Fab fragment | | Descriptor: | 1E03 Fab fragment heavy chain, 1E03 Fab fragment light chain, SULFATE ION | | Authors: | Snir, O, Chen, X, Gidoni, M, du Pre, M.F, Zhao, Y, Steinsbo, O, Lundin, K.E, Yaari, G, Sollid, L.M. | | Deposit date: | 2016-03-03 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stereotyped antibody responses target posttranslationally modified gluten in celiac disease.
JCI Insight, 2, 2017
|
|
4KNB
 
 | | C-Met in complex with OSI ligand | | Descriptor: | 7-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[3,2-c]pyridin-6-amine, GAMMA-BUTYROLACTONE, Hepatocyte growth factor receptor | | Authors: | Wang, J, Steinig, A.G, Li, A.H, Chen, X, Dong, H, Ferraro, C, Jin, M, Kadalbajoo, M, Kleinberg, A, Stolz, K.M, Tavares-Greco, P.A, Wang, T, Albertella, M.R, Peng, Y, Crew, L, Kahler, J. | | Deposit date: | 2013-05-09 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel 6-aminofuro[3,2-c]pyridines as potent, orally efficacious inhibitors of cMET and RON kinases.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3HRN
 
 | | crystal structure of a C-terminal coiled coil domain of Transient receptor potential (TRP) channel subfamily P member 2 (TRPP2, polycystic kidney disease 2) | | Descriptor: | Transient receptor potential (TRP) channel subfamily P member 2 (TRPP2) | | Authors: | Yu, Y, Ulbrich, M.H, Li, M.-H, Buraei, Z, Chen, X.-Z, Ong, A.C.M, Tong, L, Isacoff, E.Y, Yang, J. | | Deposit date: | 2009-06-09 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and molecular basis of the assembly of the TRPP2/PKD1 complex.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4IFC
 
 | | Crystal Structure of ADP-bound Human PRPF4B kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | Authors: | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | Deposit date: | 2012-12-14 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
4IJP
 
 | | Crystal Structure of Human PRPF4B kinase domain in complex with 4-{5-[(2-Chloro-pyridin-4-ylmethyl)-carbamoyl]-thiophen-2-yl}-benzo[b]thiophene-2-carboxylic acid amine | | Descriptor: | 4-(5-{[(2-chloropyridin-4-yl)methyl]carbamoyl}thiophen-2-yl)-1-benzothiophene-2-carboxamide, SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | Authors: | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | Deposit date: | 2012-12-22 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
4ME3
 
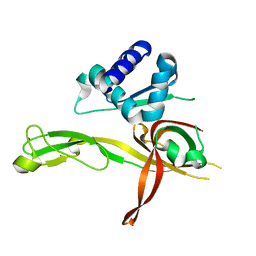 | | 1.8 Angstrom Crystal Structure of the N-terminal Domain of an Archaeal MCM | | Descriptor: | DNA replication licensing factor MCM related protein, ZINC ION | | Authors: | Fu, Y, Slaymaker, I.M, Wang, G, Chen, X.S. | | Deposit date: | 2013-08-24 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | The 1.8- angstrom Crystal Structure of the N-Terminal Domain of an Archaeal MCM as a Right-Handed Filament.
J.Mol.Biol., 426, 2014
|
|
8JQJ
 
 | | Crystal structure of carbonyl reductase SSCR mutant 1 from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
4ICK
 
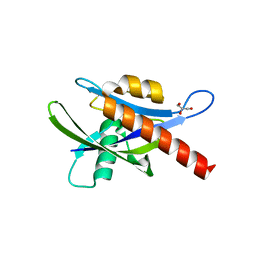 | | Crystal structure of human AP4A hydrolase E58A mutant | | Descriptor: | Bis(5'-nucleosyl)-tetraphosphatase [asymmetrical], GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ge, H, Chen, X. | | Deposit date: | 2012-12-10 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of wild-type and mutant human Ap4A hydrolase.
Biochem.Biophys.Res.Commun., 432, 2013
|
|
4IJX
 
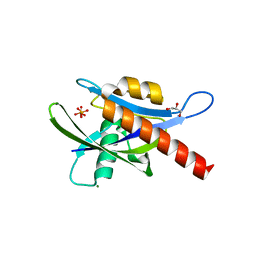 | |
6CKK
 
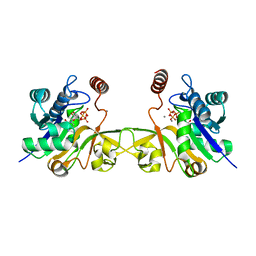 | |
6CXC
 
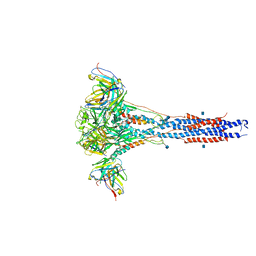 | | 3.9A Cryo-EM structure of murine antibody bound at a novel epitope of respiratory syncytial virus fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, Envelope glycoprotein chimera, ... | | Authors: | Xie, Q, Wang, Z, Chen, X, Ni, F, Ma, J, Wang, Q. | | Deposit date: | 2018-04-02 | | Release date: | 2019-07-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure basis of neutralization by a novel site II/IV antibody against respiratory syncytial virus fusion protein.
Plos One, 14, 2019
|
|
5IG7
 
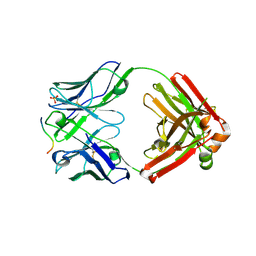 | | Crystal structure of anti-gliadin 1002-1E01 Fab fragment in complex of peptide PLQPQQPFP | | Descriptor: | 1E01 Fab fragment heavy chain, 1E01 Fab fragment light chain, SULFATE ION, ... | | Authors: | Snir, O, Chen, X, Gidoni, M, du Pre, M.F, Zhao, Y, Steinsbo, O, Lundin, K.E, Yaari, G, Sollid, L.M. | | Deposit date: | 2016-02-27 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Stereotyped antibody responses target posttranslationally modified gluten in celiac disease.
JCI Insight, 2, 2017
|
|
6BVH
 
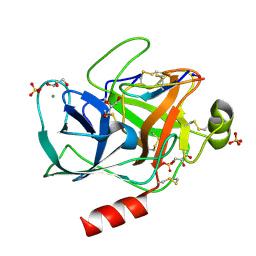 | | Trypsin complexed with a modified sunflower trypsin inhibitor, SFTI-TCTR(N12,N14) | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Riley, B.T, Chen, X. | | Deposit date: | 2017-12-13 | | Release date: | 2018-12-19 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | Potent, multi-target serine protease inhibition achieved by a simplified beta-sheet motif.
PLoS ONE, 14, 2019
|
|
5IJK
 
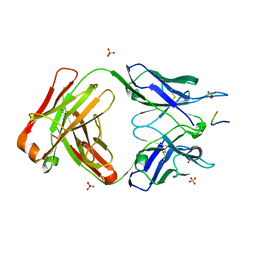 | | Crystal structure of anti-gliadin 1002-1E03 Fab fragment in complex of peptide PLQPEQPFP | | Descriptor: | 1E03 Fab fragment heavy chain, 1E03 Fab fragment light chain, SULFATE ION, ... | | Authors: | Snir, O, Chen, X, Gidoni, M, du Pre, M.F, Zhao, Y, Steinsbo, O, Lundin, K.E, Yaari, G, Sollid, L.M. | | Deposit date: | 2016-03-02 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stereotyped antibody responses target posttranslationally modified gluten in celiac disease.
JCI Insight, 2, 2017
|
|
