6LF0
 
 | | Structure of FEM1C | | Descriptor: | Protein fem-1 homolog C, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LBN
 
 | | Structure of SIL1-bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Nucleotide exchange factor SIL1 | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LEY
 
 | | Structure of Sil1G bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Nucleotide exchange factor SIL1 | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6V0V
 
 | | Cryo-EM structure of mouse WT RAG1/2 NFC complex (DNA0) | | Descriptor: | CALCIUM ION, DNA (30-MER), V(D)J recombination-activating protein 1, ... | | Authors: | Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2019-11-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OET
 
 | | Cryo-EM structure of mouse RAG1/2 STC complex | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (39-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | How mouse RAG recombinase avoids DNA transposition.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4XKC
 
 | |
4XKB
 
 | | Crystal Structure of GENOMES UNCOUPLED 4 (GUN4) in Complex with Deuteroporphyrin IX | | Descriptor: | 3,3'-(3,7,12,17-tetramethylporphyrin-2,18-diyl)dipropanoic acid, Ycf53-like protein | | Authors: | Chen, X, Pu, H, Liu, L. | | Deposit date: | 2015-01-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal Structures of GUN4 in Complex with Porphyrins.
Mol Plant, 8, 2015
|
|
6OEM
 
 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA0) | | Descriptor: | DNA (46-MER), DNA (57-MER), High mobility group protein B1, ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEP
 
 | | Cryo-EM structure of mouse RAG1/2 12RSS-NFC/23RSS-PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7XMN
 
 | | Structure of SARS-CoV-2 ORF8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltodextrin-binding protein, ... | | Authors: | Chen, X, Xu, W. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycosylated, Lipid-Binding, CDR-Like Domains of SARS-CoV-2 ORF8 Indicate Unique Sites of Immune Regulation.
Microbiol Spectr, 11, 2023
|
|
1J51
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME P450CAM MUTANT (F87W/Y96F/V247L/C334A) WITH 1,3,5-TRICHLOROBENZENE | | Descriptor: | 1,3,5-TRICHLORO-BENZENE, CYTOCHROME P450CAM, POTASSIUM ION, ... | | Authors: | Chen, X, Christopher, A, Jones, J, Guo, Q, Xu, F, Cao, R, Wong, L.L, Rao, Z. | | Deposit date: | 2002-01-05 | | Release date: | 2002-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the F87W/Y96F/V247L mutant of cytochrome P-450cam with 1,3,5-trichlorobenzene bound and further protein engineering for the oxidation of pentachlorobenzene and hexachlorobenzene
J.BIOL.CHEM., 277, 2002
|
|
3VPM
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human ribonucleotide reductase M2 subunit
To be Published
|
|
3VPO
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
7BTK
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA01 | | Descriptor: | 4-[[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-2H-indol-1-yl]methyl]benzoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Liu, Q.M, Gao, Y, Yuan, R, Guo, Y. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7BRS
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA02 | | Descriptor: | 8-[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-indol-1-ium-1-yl]octanoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Gao, Y, Yuan, R, Guo, Y. | | Deposit date: | 2020-03-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6OEN
 
 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3VPN
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
1JX6
 
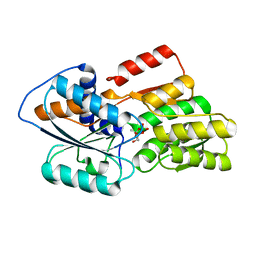 | | CRYSTAL STRUCTURE OF LUXP FROM VIBRIO HARVEYI COMPLEXED WITH AUTOINDUCER-2 | | Descriptor: | 3A-METHYL-5,6-DIHYDRO-FURO[2,3-D][1,3,2]DIOXABOROLE-2,2,6,6A-TETRAOL, CALCIUM ION, LUXP PROTEIN | | Authors: | Chen, X, Schauder, S, Potier, N, Van Dorsselaer, A, Pelczer, I, BassleR, B.L, Hughson, F.M. | | Deposit date: | 2001-09-05 | | Release date: | 2002-02-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural identification of a bacterial quorum-sensing signal containing boron.
Nature, 415, 2002
|
|
6WJN
 
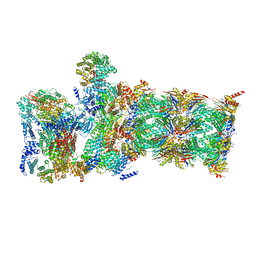 | |
2Z59
 
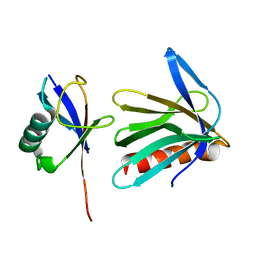 | | Complex Structures of Mouse Rpn13 (22-130aa) and ubiquitin | | Descriptor: | Protein ADRM1, Ubiquitin | | Authors: | Chen, X, Schreiner, P, Groll, M, Walters, K.J. | | Deposit date: | 2007-07-01 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction.
Nature, 453, 2008
|
|
1KIL
 
 | | Three-dimensional structure of the complexin/SNARE complex | | Descriptor: | Complexin I SNARE-complex binding region, MAGNESIUM ION, SNAP-25 C-terminal SNARE motif, ... | | Authors: | Chen, X, Tomchick, D, Kovrigin, E, Arac, D, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-12-03 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the complexin/SNARE complex.
Neuron, 33, 2002
|
|
1EKD
 
 | | NMR AND MOLECULAR MODELING REVEAL THAT DIFFERENT HYDROGEN BONDING PATTERNS ARE POSSIBLE FOR GU PAIRS: ONE HYDROGEN BOND FOR EACH GU PAIR IN R(GGCGUGCC)2 AND TWO FOR EACH GU PAIR IN R(GAGUGCUC)2 | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*GP*CP*C)-3') | | Authors: | Chen, X, McDowell, J.A, Kierzek, R, Krugh, T.R, Turner, D.H. | | Deposit date: | 2000-03-07 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2).
Biochemistry, 39, 2000
|
|
4RWT
 
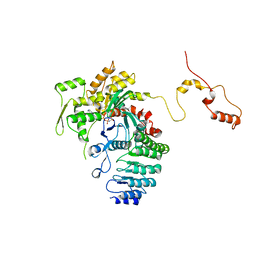 | | Structure of actin-Lmod complex | | Descriptor: | Actin-5C, Leiomodin-2, MAGNESIUM ION, ... | | Authors: | Chen, X, Ni, F, Wang, Q. | | Deposit date: | 2014-12-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Mechanisms of leiomodin 2-mediated regulation of actin filament in muscle cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5ZXF
 
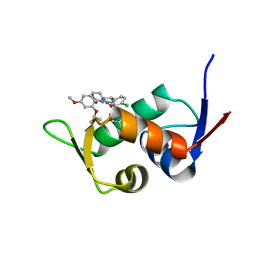 | | The 1.25A Crystal structure of His6-tagged Mdm2 in complex with nutlin-3a | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Cheng, X.Y, Su, Z.D, Pi, N, Cao, C.Z, Zhao, Z.T, Zhou, J.J, Chen, R, Kuang, Z.K, Huang, Y.Q. | | Deposit date: | 2018-05-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The 1.25A Crystal structure of His6-tagged Mdm2 in complex with nutlin-3a
To Be Published
|
|
6WJD
 
 | |
