5T0G
 
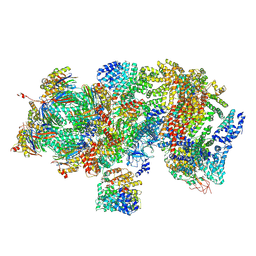 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2016-11-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0H
 
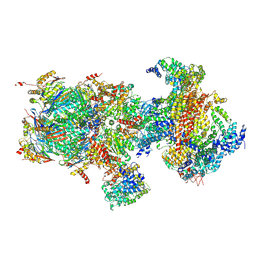 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2016-11-30 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5VOU
 
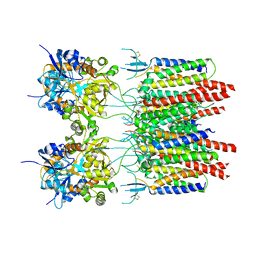 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Chen, S, Zhao, Y, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
1F5T
 
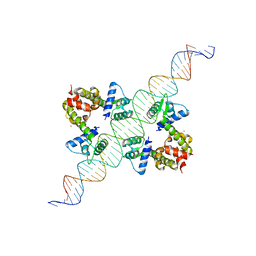 | | DIPHTHERIA TOX REPRESSOR (C102D MUTANT) COMPLEXED WITH NICKEL AND DTXR CONSENSUS BINDING SEQUENCE | | Descriptor: | 43MER DNA CONTAINING DXTR CONSENSUS BINDING SEQUENCE, DIPHTHERIA TOXIN REPRESSOR, NICKEL (II) ION | | Authors: | Chen, S, White, A, Love, J, Murphy, J.R, Ringe, D. | | Deposit date: | 2000-06-15 | | Release date: | 2000-09-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Methyl groups of thymine bases are important for nucleic acid recognition by DtxR.
Biochemistry, 39, 2000
|
|
7F5F
 
 | | SARS-CoV-2 ORF8 S84 | | Descriptor: | CALCIUM ION, ORF8 protein | | Authors: | Chen, S, Zhou, Z, Chen, X. | | Deposit date: | 2021-06-22 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structures of Bat and Human Coronavirus ORF8 Protein Ig-Like Domain Provide Insights Into the Diversity of Immune Responses.
Front Immunol, 12, 2021
|
|
1S3M
 
 | | Structural and Functional Characterization of a Novel Archaeal Phosphodiesterase | | Descriptor: | Hypothetical protein MJ0936, NICKEL (II) ION | | Authors: | Chen, S, Busso, D, Yakunin, A.F, Kuznetsova, E, Proudfoot, M, Jancrick, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii
J.Biol.Chem., 279, 2004
|
|
1S3N
 
 | | Structural and Functional Characterization of a Novel Archaeal Phosphodiesterase | | Descriptor: | Hypothetical protein MJ0936, MANGANESE (II) ION | | Authors: | Chen, S, Busso, D, Yakunin, A.F, Kuznetsova, E, Proudfoot, M, Jancrick, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii
J.Biol.Chem., 279, 2004
|
|
1S3L
 
 | | Structural and Functional Characterization of a Novel Archaeal Phosphodiesterase | | Descriptor: | Hypothetical protein MJ0936, PHOSPHATE ION, UNKNOWN ATOM OR ION | | Authors: | Chen, S, Busso, D, Yakunin, A.F, Kuznetsova, E, Proudfoot, M, Jancrick, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii
J.Biol.Chem., 279, 2004
|
|
1SBQ
 
 | | Crystal Structure of methenyltetrahydrofolate synthetase from Mycoplasma pneumoniae at 2.2 resolution | | Descriptor: | 5,10-Methenyltetrahydrofolate synthetase homolog, SULFATE ION | | Authors: | Chen, S, Shin, D.H, Pufan, R, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-02-10 | | Release date: | 2004-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of methenyltetrahydrofolate synthetase from Mycoplasma pneumoniae (GI: 13508087) at 2.2 A resolution
Proteins, 56, 2004
|
|
1N0F
 
 | | CRYSTAL STRUCTURE OF A CELL DIVISION AND CELL WALL BIOSYNTHESIS PROTEIN UPF0040 FROM MYCOPLASMA PNEUMONIAE: INDICATION OF A NOVEL FOLD WITH A POSSIBLE NEW CONSERVED SEQUENCE MOTIF | | Descriptor: | Protein mraZ | | Authors: | Chen, S, Jancrick, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-10-13 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a protein associated with cell division from Mycoplasma pneumoniae (GI: 13508053): a novel fold with a conserved sequence motif.
Proteins, 55, 2004
|
|
1N0G
 
 | | Crystal Structure of A Cell Division and Cell Wall Biosynthesis Protein UPF0040 from Mycoplasma pneumoniae: Indication of A Novel Fold with A Possible New Conserved Sequence Motif | | Descriptor: | Protein mraZ | | Authors: | Chen, S, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-10-13 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a protein associated with cell division from Mycoplasma pneumoniae (GI: 13508053): a novel fold with a conserved sequence motif.
Proteins, 55, 2004
|
|
1N0E
 
 | | CRYSTAL STRUCTURE OF A CELL DIVISION AND CELL WALL BIOSYNTHESIS PROTEIN UPF0040 FROM MYCOPLASMA PNEUMONIAE: INDICATION OF A NOVEL FOLD WITH A POSSIBLE NEW CONSERVED SEQUENCE MOTIF | | Descriptor: | Protein mraZ | | Authors: | Chen, S, Jancrick, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-10-13 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a protein associated with cell division from Mycoplasma pneumoniae (GI: 13508053): a novel fold with a conserved sequence motif.
Proteins, 55, 2004
|
|
6M3M
 
 | |
1U3F
 
 | | Structural and Functional Characterization of a 5,10-Methenyltetrahydrofolate Synthetase from Mycoplasma pneumoniae (GI: 13508087) | | Descriptor: | 5,10-Methenyltetrahydrofolate Synthetase, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chen, S, Yakunin, A.F, Proudfoot, M, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-07-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of methenyltetrahydrofolate synthetase from Mycoplasma pneumoniae (GI: 13508087) at 2.2 A resolution
Proteins, 56, 2004
|
|
1U3G
 
 | | Structural and Functional Characterization of a 5,10-Methenyltetrahydrofolate Synthetase from Mycoplasma pneumoniae (GI: 13508087) | | Descriptor: | 5,10-Methenyltetrahydrofolate Synthetase, 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, S, Yakunin, A.F, Proudfoot, M, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-07-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of methenyltetrahydrofolate synthetase from Mycoplasma pneumoniae (GI: 13508087) at 2.2 A resolution
Proteins, 56, 2004
|
|
7CR5
 
 | |
5DAH
 
 | |
4JBW
 
 | | Crystal structure of E. coli maltose transporter MalFGK2 in complex with its regulatory protein EIIAglc | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Glucose-specific phosphotransferase enzyme IIA component, Maltose transport system permease protein MalF, ... | | Authors: | Chen, S, Oldham, M.L, Davidson, A.L, Chen, J. | | Deposit date: | 2013-02-20 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.913 Å) | | Cite: | Carbon catabolite repression of the maltose transporter revealed by X-ray crystallography.
Nature, 499, 2013
|
|
7DE1
 
 | |
4JLW
 
 | | Crystal structure of formaldehyde dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | Glutathione-independent formaldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Chen, S, Liao, Y.P, Wang, D.L, Wang, S, Ding, J.F, Wang, Y.M, Cai, L.J, Ran, X.Y, Zhu, H.X. | | Deposit date: | 2013-03-13 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of formaldehyde dehydrogenase from Pseudomonas aeruginosa: the binary complex with the cofactor NAD+.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5DAG
 
 | |
8K1V
 
 | |
8K1Q
 
 | |
8K1Z
 
 | |
8K1J
 
 | |
