1S9W
 
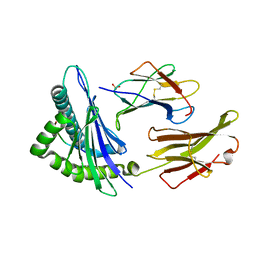 | | Crystal Structure Analysis of NY-ESO-1 epitope, SLLMWITQC, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
5BMV
 
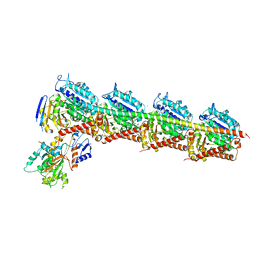 | | CRYSTAL STRUCTURE OF TUBULIN-STATHMIN-TTL-Vinblastine COMPLEX | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Chen, Q, Zhang, R. | | Deposit date: | 2015-05-23 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules.
Mol.Pharmacol., 89, 2016
|
|
2EXX
 
 | | Crystal structure of HSCARG from Homo sapiens in complex with NADP | | Descriptor: | GLYCEROL, HSCARG protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dai, X, Chen, Q, Yao, D, Liang, Y, Dong, Y, Gu, X, Zheng, X, Luo, M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Restructuring of the dinucleotide-binding fold in an NADP(H) sensor protein.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
3WL0
 
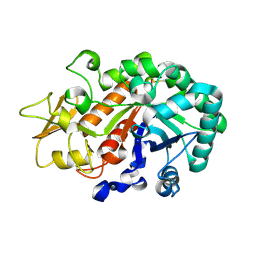 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain E148A mutant in complex with a(GlcNAc)2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WKZ
 
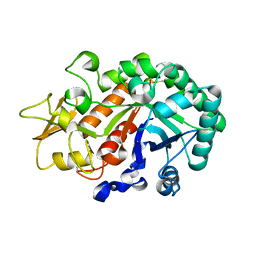 | | Crystal Structure of the Ostrinia furnacalis Group I Chitinase catalytic domain E148Q mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WL1
 
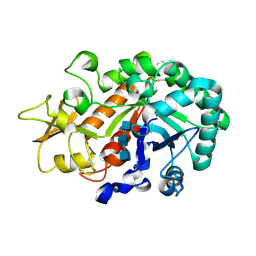 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain in complex with reaction products (GlcNAc)2,3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5T38
 
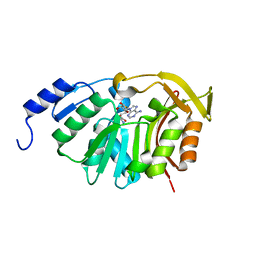 | | Crystal Structure of the N-terminal domain of EvdMO1 with SAH bound | | Descriptor: | EvdMO1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McCulloch, K.M, Berndt, S, Yamakawa, I, Chen, Q, Loukachevitch, L.V, Starbird, C, Perry, N.A, Iverson, T.M. | | Deposit date: | 2016-08-25 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1502 Å) | | Cite: | The Structure of the Bifunctional Everninomicin Biosynthetic Enzyme EvdMO1 Suggests Independent Activity of the Fused Methyltransferase-Oxidase Domains.
Biochemistry, 57, 2018
|
|
5GT9
 
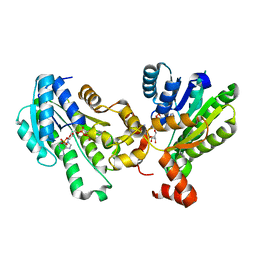 | | The X-ray structure of 7beta-hydroxysteroid dehydrogenase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, short chain dehydrogenase/reductase family protein | | Authors: | Wang, F, Wang, R, Lv, Z, Chen, Q, Huo, X. | | Deposit date: | 2016-08-19 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of NADP(+)-bound 7 beta-hydroxysteroid dehydrogenase reveals two cofactor-binding modes
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4XBS
 
 | | 2-deoxyribose-5-phosphate aldolase mutant - E78K | | Descriptor: | Deoxyribose-phosphate aldolase | | Authors: | Jiao, X.-C, Pan, J, Xu, G.-C, Kong, X.-D, Chen, Q, Zhang, Z.-J, Xu, J.-H. | | Deposit date: | 2014-12-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Efficient synthesis of a statin precursor in high space-time yield by a new aldehyde-tolerant aldolase identified from Lactobacillus brevis
Catalysis Science And Technology, 5, 2015
|
|
4XBK
 
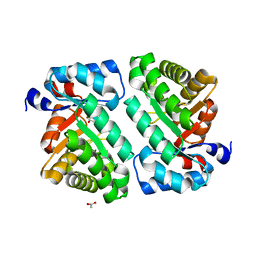 | | 2-deoxyribose-5-phosphate aldolase from Lactobacillus brevis | | Descriptor: | ACETIC ACID, Deoxyribose-phosphate aldolase | | Authors: | Jiao, X.-C, Pan, J, Xu, G.-C, Kong, X.-D, Chen, Q, Zhang, Z.-J, Xu, J.-H. | | Deposit date: | 2014-12-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Efficient synthesis of a statin precursor in high space-time yield by a new aldehyde-tolerant aldolase identified from Lactobacillus brevis
Catalysis Science And Technology, 5, 2015
|
|
4B02
 
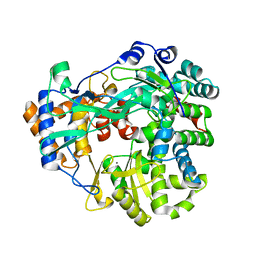 | | The C-terminal Priming Domain is Strongly Associated with the Main Body of Bacteriophage phi6 RNA-Dependent RNA Polymerase | | Descriptor: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Sarin, L.P, Wright, S, Chen, Q, Degerth, L.H, Stuart, D.I, Grimes, J.M, Bamford, D.H, Poranen, M.M. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The C-Terminal Priming Domain is Strongly Associated with the Main Body of Bacteriophage Phi6 RNA-Dependent RNA Polymerase.
Virology, 432, 2012
|
|
1NT2
 
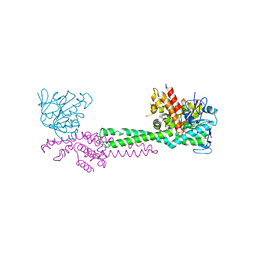 | | CRYSTAL STRUCTURE OF FIBRILLARIN/NOP5P COMPLEX | | Descriptor: | Fibrillarin-like pre-rRNA processing protein, S-ADENOSYLMETHIONINE, conserved hypothetical protein | | Authors: | Aittaleb, M, Rashid, R, Chen, Q, Palmer, J.R, Daniels, C.J, Li, H. | | Deposit date: | 2003-01-28 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and function of archaeal box C/D sRNP core proteins.
Nat.Struct.Biol., 10, 2003
|
|
4J0J
 
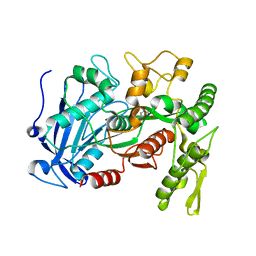 | | Tannin acyl hydrolase in complex with ethyl 3,5-dihydroxybenzoate | | Descriptor: | Tannase, ethyl 3,5-dihydroxybenzoate | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-31 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0G
 
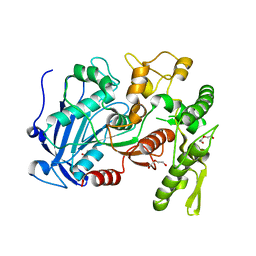 | | Tannin acyl hydrolase (mercury derivative) | | Descriptor: | DI(HYDROXYETHYL)ETHER, MERCURY (II) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-30 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0C
 
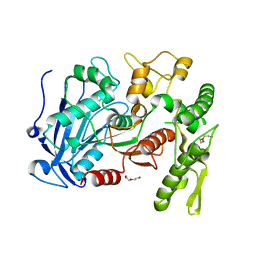 | | tannin acyl hydrolase from Lactobacillus plantarum (native structure) | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Tannase | | Authors: | Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-30 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0K
 
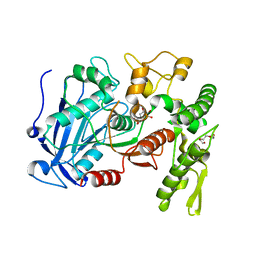 | | Tannin acyl hydrolase in complex with ethyl gallate | | Descriptor: | DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-31 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0D
 
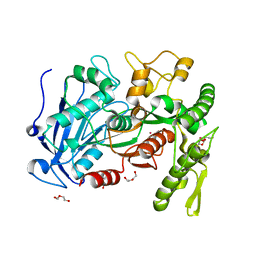 | | tannin acyl hydrolase from Lactobacillus plantarum (Cadmium) | | Descriptor: | CADMIUM ION, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-30 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0H
 
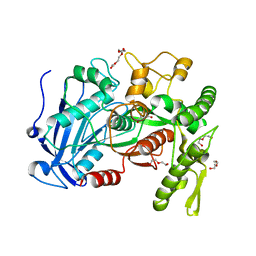 | | Tannin acyl hydrolase in complex with gallic acid | | Descriptor: | 3,4,5-trihydroxybenzoic acid, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-30 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0I
 
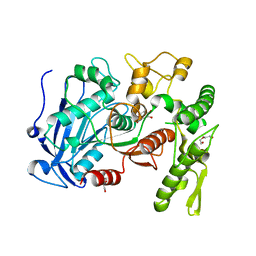 | | Tannin acyl hydrolase in complex with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-30 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4JUI
 
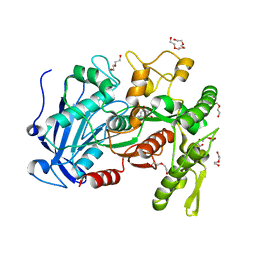 | | crystal structure of tannase from from Lactobacillus plantarum | | Descriptor: | DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, McKinstry, W.J, Chen, Q. | | Deposit date: | 2013-03-24 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4QXW
 
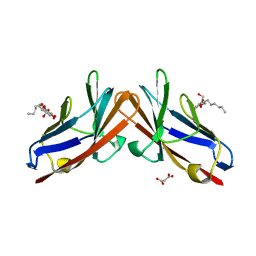 | | Crystal structure of the human CEACAM1 membrane distal amino terminal (N)-domain | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID, octyl beta-D-glucopyranoside | | Authors: | Huang, Y.H, Gandhi, A.K, Russell, A, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2014-07-22 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
8F4V
 
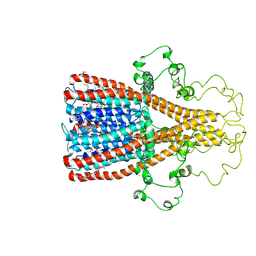 | | Alpha7 nicotinic acetylcholine receptor intracellular and transmembrane domains bound to ivermectin in a desensitized state | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, Neuronal acetylcholine receptor subunit alpha-7 | | Authors: | Bondarenko, V, Chen, Q, Tang, P. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Elucidation of Ivermectin Binding to alpha 7nAChR and the Induced Channel Desensitization.
Acs Chem Neurosci, 14, 2023
|
|
7XU2
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU0
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-211 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XTZ
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
