3J7M
 
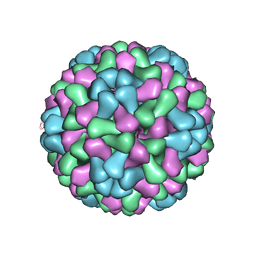 | | Virus model of brome mosaic virus (first half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
4IMO
 
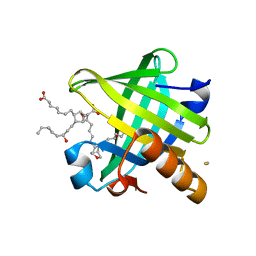 | | Crystal structure of wild type human Lipocalin PGDS in complex with substrate analog U44069 | | Descriptor: | (5E)-7-{(1R,4S,5S,6R)-5-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-6-yl}hept-5-enoic acid, Lipocalin-type prostaglandin-D synthase, THIOCYANATE ION | | Authors: | Lim, S.M, Chen, D, Teo, H, Roos, A, Nyman, T, Tresaugues, L, Pervushin, K, Nordlund, P. | | Deposit date: | 2013-01-03 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and dynamic insights into substrate binding and catalysis of human lipocalin prostaglandin D synthase.
J.Lipid Res., 54, 2013
|
|
4IMN
 
 | | Crystal structure of wild type human Lipocalin PGDS bound with PEG MME 2000 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Lipocalin-type prostaglandin-D synthase | | Authors: | Lim, S.M, Chen, D, Teo, H, Roos, A, Nyman, T, Tresaugues, L, Pervushin, K, Nordlund, P. | | Deposit date: | 2013-01-03 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and dynamic insights into substrate binding and catalysis of human lipocalin prostaglandin D synthase.
J.Lipid Res., 54, 2013
|
|
4JDN
 
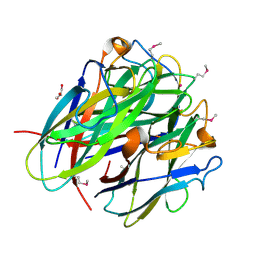 | | Secreted Chlamydial Protein PGP3, C-terminal Domain | | Descriptor: | GLYCEROL, POTASSIUM ION, Virulence plasmid protein pGP3-D | | Authors: | Galaleldeen, A, Taylor, A.B, Chen, D, Holloway, S.P, Zhong, G, Hart, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-06-05 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Chlamydia trachomatis immunodominant antigen Pgp3.
J.Biol.Chem., 288, 2013
|
|
4JDM
 
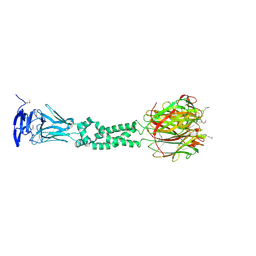 | | Secreted Chlamydial Protein PGP3, full-length | | Descriptor: | Virulence plasmid protein pGP3-D | | Authors: | Galaleldeen, A, Taylor, A.B, Chen, D, Holloway, S.P, Zhong, G, Hart, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-06-05 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Chlamydia trachomatis immunodominant antigen Pgp3.
J.Biol.Chem., 288, 2013
|
|
4JDO
 
 | | Secreted chlamydial protein pgp3, coiled-coil deletion | | Descriptor: | SODIUM ION, Virulence plasmid protein pGP3-D | | Authors: | Galaleldeen, A, Taylor, A.B, Chen, D, Holloway, S.P, Zhong, G, Hart, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Chlamydia trachomatis immunodominant antigen Pgp3.
J.Biol.Chem., 288, 2013
|
|
5GAI
 
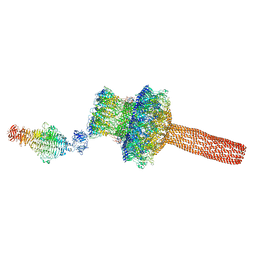 | | Probabilistic Structural Models of Mature P22 Bacteriophage Portal, Hub, and Tailspike proteins | | Descriptor: | Peptidoglycan hydrolase gp4, Portal protein, Tail fiber protein | | Authors: | Pintilie, G, Chen, D.H, Haase-Pettingell, C.A, King, J.A, Chiu, W. | | Deposit date: | 2015-12-01 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Resolution and Probabilistic Models of Components in CryoEM Maps of Mature P22 Bacteriophage.
Biophys.J., 110, 2016
|
|
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | Descriptor: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | Authors: | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
6K05
 
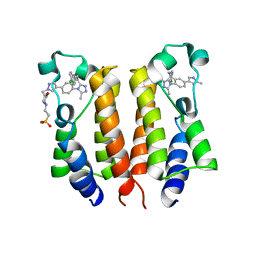 | | Crystal structure of BRD2(BD1)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
6AHZ
 
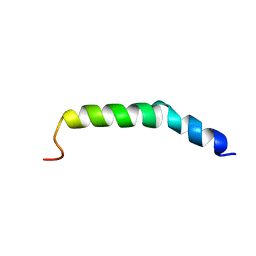 | | The NMR Structure of the Polysialyltranseferase Domain (PSTD) in Polysialyltransferase ST8siaIV | | Descriptor: | CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase | | Authors: | Liu, X.H, Lu, B, Peng, L.X, Liao, S.M, Zhou, F, Chen, D, Lu, Z.L, Zhou, G.P, Huang, R.B. | | Deposit date: | 2018-08-21 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Inhibition of Polysialyltranseferase ST8SiaIV Through Heparin Binding to Polysialyltransferase Domain (PSTD).
Med Chem, 15, 2019
|
|
6K04
 
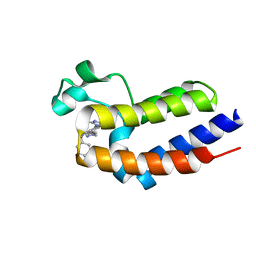 | | Crystal structure of BRD2(BD2)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
5YFM
 
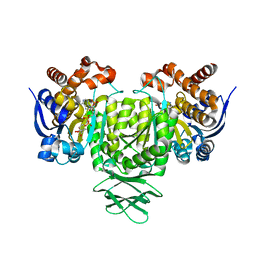 | |
5YFN
 
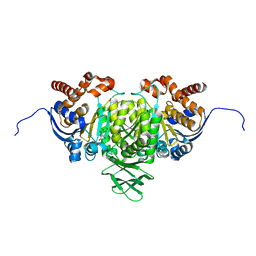 | | Human isocitrate dehydrogenase 1 bound with isocitrate | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase [NADP] cytoplasmic, MAGNESIUM ION, ... | | Authors: | Nordlund, P, Chen, D, Jansson, A, Larsson, A. | | Deposit date: | 2017-09-21 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human isocitrate dehydrogenase 1 bound with isocitrate
To Be Published
|
|
7JMN
 
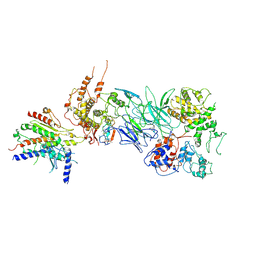 | | Tail module of Mediator complex | | Descriptor: | MED15, Mediator of RNA polymerase II transcription subunit 14, Mediator of RNA polymerase II transcription subunit 16, ... | | Authors: | Zhang, H.Q, Chen, D.C. | | Deposit date: | 2020-08-02 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Mediator structure and conformation change.
Mol.Cell, 81, 2021
|
|
5Y22
 
 | | NMR-Based Model of the 22 Amino Acid Peptide in Polysialyltransferase Domain (PSTD) of the Polysialyltransferase ST8Sia IV | | Descriptor: | 22AA-PSTD peptide | | Authors: | Lu, B, Liao, S.M, Huang, J.M, Lu, Z.L, Chen, D, Liu, X.H, Zhou, G.P, Huang, R.B. | | Deposit date: | 2017-07-23 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR-Based Model of the 22 Amino Acid Peptide in Polysialyltransferase Domain (PSTD) of the Polysialyltransferase ST8Sia IV
To Be Published
|
|
7DLZ
 
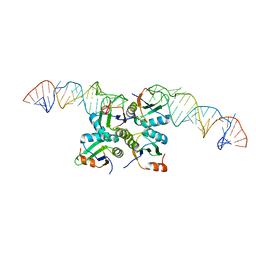 | | Crystal Structure of Methyltransferase Ribozyme | | Descriptor: | RNA (45-MER), U1 small nuclear ribonucleoprotein A | | Authors: | Gan, J.H, Gao, Y.Q, Jiang, H.Y, Chen, D.R, Murchie, A.I.H. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences
Nat Catal, 4, 2021
|
|
7DWH
 
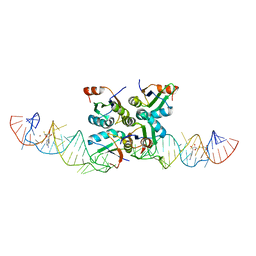 | | Complex structure of SAM-dependent methyltransferase ribozyme | | Descriptor: | COPPER (II) ION, RNA (45-MER), S-ADENOSYLMETHIONINE, ... | | Authors: | Jiang, H.Y, Gao, Y.Q, Chen, D.R, Murchie, A. | | Deposit date: | 2021-01-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences
Nat Catal, 4, 2021
|
|
7VH6
 
 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the active state (pH 6.0, BeF3-, conformation 1, C1 symmetry) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, BERYLLIUM TRIFLUORIDE ION, Plasma membrane ATPase 1 | | Authors: | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-24 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
7VH5
 
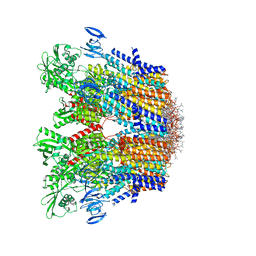 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the autoinhibited state (pH 7.4, C1 symmetry) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Plasma membrane ATPase 1, SPHINGOSINE | | Authors: | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-24 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
4J3J
 
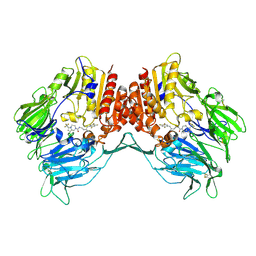 | | Crystal Structure of DPP-IV with Compound C3 | | Descriptor: | Dipeptidyl peptidase 4, N-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butyl]-6-(trifluoromethyl)-3,4-dihydropyrrolo[1,2-a]pyrazine-2(1H)-carboxamide | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2013-02-05 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 4-(2,4,5-trifluorophenyl)butane-1,3-diamines as dipeptidyl peptidase IV inhibitors
Chemmedchem, 8, 2013
|
|
5Y3U
 
 | | NMR-Based Model of the 22 Amino Acid Peptide in Polysialyltransferase Domain (PSTD) of the Polysialyltransferase ST8Sia IV in the Presence of Polysialic Acid (PolySia) | | Descriptor: | PSTD-22AA-PolySia | | Authors: | Liao, S.M, Liu, X.H, Lu, B, Peng, L.X, Chen, D, Huang, R.B, Zhou, G.P. | | Deposit date: | 2017-07-31 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR-Based Model of the 22 Amino Acid Peptide in Polysialyltransferase Domain (PSTD) of the Polysialyltransferase ST8Sia IV in the Presence of Polysialic Acid (PolySia)
To Be Published
|
|
