6J02
 
 | | Crystal structure of the SRCR domain of mouse SCARA1 | | Descriptor: | CALCIUM ION, Macrophage scavenger receptor types I and II | | Authors: | Cheng, C, Hu, Z. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | The scavenger receptor SCARA1 (CD204) recognizes dead cells through spectrin.
J.Biol.Chem., 294, 2019
|
|
4D4E
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARMADILLO REPEAT PROTEIN ARM00016, GLYCEROL | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4D49
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARGININE, ARMADILLO REPEAT PROTEIN ARM00027, POLY ARG DECAPEPTIDE | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4RUD
 
 | | Crystal structure of a three finger toxin | | Descriptor: | Three-finger toxin 3b, ZINC ION | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2014-11-18 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fulditoxin, representing a new class of dimeric snake toxins, defines novel pharmacology at nicotinic ACh receptors.
Br.J.Pharmacol., 177, 2020
|
|
4PLS
 
 | | Crystal Structures of Designed Armadillo Repeat Proteins: Implications of Construct Design and Crystallization Conditions on Overall Structure. | | Descriptor: | ACETATE ION, Arm00010, CALCIUM ION | | Authors: | Reichen, C, Madhurantakam, C, Plueckthun, A, Mittl, P.R. | | Deposit date: | 2014-05-19 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of Designed Armadillo Repeat Proteins: Implications of Construct Design and Crystallization Conditions on Overall Structure.
Protein Sci., 23, 2014
|
|
4PLR
 
 | | Crystal Structures of Designed Armadillo Repeat Proteins: Implications of Construct Design and Crystallization Conditions on Overall Structure. | | Descriptor: | Arm00008, CALCIUM ION | | Authors: | Reichen, C, Madhurantakam, C, Plueckthun, A, Mittl, P. | | Deposit date: | 2014-05-19 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of designed armadillo repeat proteins: Implications of construct design and crystallization conditions on overall structure.
Protein Sci., 23, 2014
|
|
7C28
 
 | | Unusual quaternary structure of a homodimeric synergistic toxin from mamba snake venom | | Descriptor: | SULFATE ION, Synergistic-type venom protein S2C4 | | Authors: | Jobichen, C, Narumi, A, Sivaraman, J, Kini, R.M. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unusual quaternary structure of a homodimeric synergistic-type toxin from mamba snake venom defines its molecular evolution.
Biochem.J., 477, 2020
|
|
4ZQY
 
 | |
2ZQV
 
 | |
2ZQU
 
 | |
2ZR4
 
 | |
2ZR5
 
 | |
2ZQS
 
 | |
2ZR6
 
 | |
2ZQT
 
 | |
6A2V
 
 | | Crystal structure of Hcp protein | | Descriptor: | Type VI secretion system tube protein Hcp | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2018-06-13 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.588 Å) | | Cite: | Structural basis for the pathogenesis of Campylobacter jejuni Hcp1, a structural and effector protein of the Type VI Secretion System.
FEBS J., 285, 2018
|
|
1A41
 
 | | TYPE 1-TOPOISOMERASE CATALYTIC FRAGMENT FROM VACCINIA VIRUS | | Descriptor: | SULFATE ION, TOPOISOMERASE I | | Authors: | Cheng, C, Kussie, P, Pavletich, N, Shuman, S. | | Deposit date: | 1998-02-10 | | Release date: | 1999-06-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases.
Cell(Cambridge,Mass.), 92, 1998
|
|
3UT8
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | IODIDE ION, Putative uncharacterized protein | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3UT7
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3UT4
 
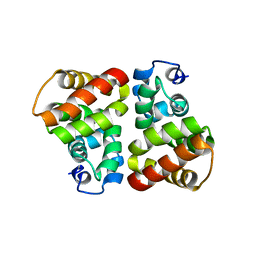 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | Putative uncharacterized protein | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
8JB0
 
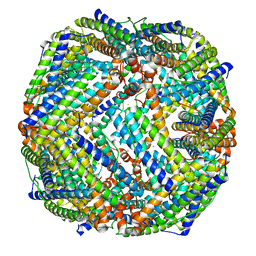 | |
8JAX
 
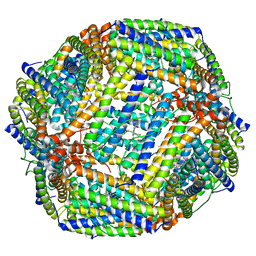 | |
5XX9
 
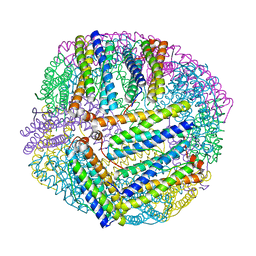 | |
5XWR
 
 | | Crystal Structure of RBBP4-peptide complex | | Descriptor: | Histone-binding protein RBBP4, MET-SER-ARG-ARG-LYS-GLN-ALA-LYS-PRO-GLN-HIS-ILE | | Authors: | Jobichen, C, Lui, B.H, Daniel, G.T, Sivaraman, J. | | Deposit date: | 2017-06-30 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XWE
 
 | | Structure of a three finger toxin from Ophiophagus hannah venom | | Descriptor: | CHLORIDE ION, GLYCEROL, Weak toxin DE-1 homolog 1, ... | | Authors: | Jobichen, C, Roy, A, Kini, R.M, Sivaraman, J. | | Deposit date: | 2017-06-29 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a three finger toxin from Ophiophagus hannah venom
To Be Published
|
|
