3SUB
 
 | |
2FN7
 
 | |
2H2Q
 
 | | Crystal structure of Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate synthase | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Senkovich, O, Schormann, N, Chattopadhyay, D. | | Deposit date: | 2006-05-19 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
1IH8
 
 | | NH3-dependent NAD+ Synthetase from Bacillus subtilis Complexed with AMP-CPP and Mg2+ ions. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) synthetase | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, DeLucas, L. | | Deposit date: | 2001-04-18 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3KJS
 
 | | Crystal Structure of T. cruzi DHFR-TS with 3 high affinity DHFR inhibitors: DQ1 inhibitor complex | | Descriptor: | 1,2-ETHANEDIOL, Dihydrofolate reductase-thymidylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2009-11-03 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis and characterization of potent inhibitors of Trypanosoma cruzi dihydrofolate reductase.
Bioorg.Med.Chem., 18, 2010
|
|
3HBB
 
 | | Structures of dihydrofolate reductase-thymidylate synthase of Trypanosoma cruzi in the folate-free state and in complex with two antifolate drugs, trimetrexate and methotrexate | | Descriptor: | 1,2-ETHANEDIOL, Dihydrofolate reductase-thymidylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of dihydrofolate reductase-thymidylate synthase of Trypanosoma cruzi in the folate-free state and in complex with two antifolate drugs, trimetrexate and methotrexate.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3LRP
 
 | |
1NX3
 
 | | Calpain Domain VI in Complex with the Inhibitor PD150606 | | Descriptor: | 3-(4-IODO-PHENYL)-2-MERCAPTO-PROPIONIC ACID, CALCIUM ION, Calcium-dependent protease, ... | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
1VSV
 
 | |
1VSU
 
 | |
4DOG
 
 | |
4DOF
 
 | |
1I1N
 
 | | HUMAN PROTEIN L-ISOASPARTATE O-METHYLTRANSFERASE WITH S-ADENOSYL HOMOCYSTEINE | | Descriptor: | PROTEIN-L-ISOASPARTATE O-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Smith, C.D, Chattopadhyay, D, Carson, M, Friedman, A.M, Skinner, M.M. | | Deposit date: | 2001-02-02 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human L-isoaspartyl-O-methyl-transferase with S-adenosyl homocysteine at 1.6-A resolution and modeling of an isoaspartyl-containing peptide at the active site.
Protein Sci., 11, 2002
|
|
3OZ7
 
 | |
4LZB
 
 | | Uracil binding pocket in Vaccinia virus uracil DNA glycosylase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the uracil complex of Vaccinia virus uracil DNA glycosylase.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3OZA
 
 | |
1RDH
 
 | |
1QCP
 
 | | CRYSTAL STRUCTURE OF THE RWJ-51084 BOVINE PANCREATIC BETA-TRYPSIN AT 1.8 A | | Descriptor: | CALCIUM ION, CYCLOPENTANECARBOXYLIC ACID [1-(BENZOTHIAZOLE-2-CARBONYL)-4-GUANIDINO-BUTYL]-AMIDE, PROTEIN (BETA-TRYPSIN PROTEIN) | | Authors: | Recacha, R, Carson, M, Costanzo, M.J, Maryanoff, B, Chattopadhyay, D. | | Deposit date: | 1999-05-10 | | Release date: | 1999-05-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the RWJ-51084-bovine pancreatic beta-trypsin complex at 1.8 A.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5JYF
 
 | |
5JY6
 
 | |
5JX8
 
 | |
5JYE
 
 | |
5JX3
 
 | | Wild type D4 in orthorhombic space group | | Descriptor: | CHLORIDE ION, GLYCEROL, Uracil-DNA glycosylase | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2016-05-12 | | Release date: | 2016-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Poxvirus uracil-DNA glycosylase-An unusual member of the family I uracil-DNA glycosylases.
Protein Sci., 25, 2016
|
|
4QC9
 
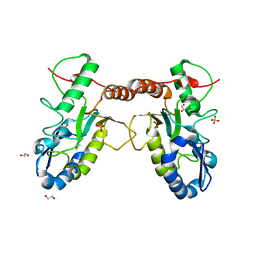 | | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uracil-DNA glycosylase | | Authors: | Sartmatova, D, Nash, T, Schormann, N, Nuth, M, Ricciardi, R, Banerjee, S, Chattopadhyay, D. | | Deposit date: | 2014-05-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.259 Å) | | Cite: | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4
To be Published
|
|
1MXF
 
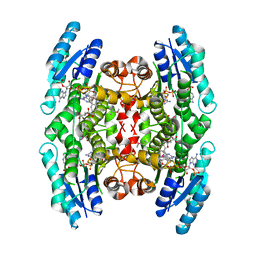 | | Crystal Structure of Inhibitor Complex of Putative Pteridine Reductase 2 (PTR2) from Trypanosoma cruzi | | Descriptor: | METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 2 | | Authors: | Schormann, N, Pal, B, Senkovich, O, Carson, M, Howard, A, Smith, C, Delucas, L, Chattopadhyay, D. | | Deposit date: | 2002-10-02 | | Release date: | 2003-10-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Trypanosoma cruzi pteridine reductase 2 in complex with a substrate and an inhibitor.
J.Struct.Biol., 152, 2005
|
|
