7UDJ
 
 | | Crystal structure of designed helical repeat protein RPB_PEW3_R4 bound to PAWx4 peptide | | Descriptor: | 4xPAW peptide, De novo designed helical repeat protein RPB_PEW3_R4 | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
2WIC
 
 | | Crystal Structures of the N-terminal Intracellular Domain of FeoB from Klebsiella Pneumoniae in GMPPNP binding state | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Hung, K.-W, Chang, Y.-W, Chen, J.-H, Chen, Y.-C, Sun, Y.-J, Hsiao, C.-D, Huang, T.-H. | | Deposit date: | 2009-05-09 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Fold, Conservation and Fe(II) Binding of the Intracellular Domain of Prokaryote Feob.
J.Struct.Biol., 170, 2010
|
|
3OS5
 
 | | SET7/9-Dnmt1 K142me1 complex | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Dnmt1, ... | | Authors: | Esteve, P.-O, Chang, Y, Samaranayake, M, Upadhyay, A.K, Horton, J.R, Feehery, G.R, Cheng, X, Pradhan, S. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7UDO
 
 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 (proteolysis fragment?), forming pseudopolymeric filaments | | Descriptor: | 1,2-ETHANEDIOL, Designed helical repeat protein (DHR) RPB_LRP2_R4, PHOSPHATE ION | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
8IQE
 
 | | Crystal structure of tetrameric K2-2 TSP | | Descriptor: | GLYCEROL, K2-VCL6 TSP | | Authors: | Ye, T.J, Huang, K.F, Tu, I.F, Lee, I.M, Chang, Y.P, Wu, S.H. | | Deposit date: | 2023-03-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Klebsiella pneumoniae K2 capsular polysaccharide degradation by a bacteriophage depolymerase does not require trimer formation.
Mbio, 15, 2024
|
|
7V84
 
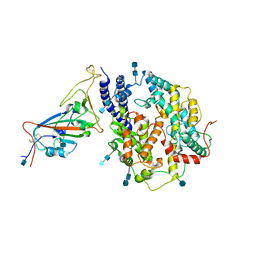 | | Local refinement of SARS-CoV-2 S-Gamma variant (P.1) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Local refinement of SARS-CoV-2 S-Gamma variant (P.1) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain
To Be Published
|
|
7V8B
 
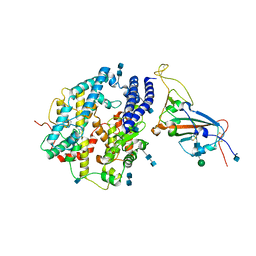 | | Local refinement of SARS-CoV-2 S-Delta variant (B.1.617.2) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Local refinement of SARS-CoV-2 S-Delta variant (B.1.617.2) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain
To Be Published
|
|
2JW8
 
 | | Solution structure of stereo-array isotope labelled (SAIL) C-terminal dimerization domain of SARS coronavirus nucleocapsid protein | | Descriptor: | Nucleocapsid protein | | Authors: | Takeda, M, Chang, C, Ikeya, T, Guntert, P, Chang, Y, Hsu, Y, Huang, T, Kainosho, M. | | Deposit date: | 2007-10-06 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method
J.Mol.Biol., 380, 2008
|
|
7V76
 
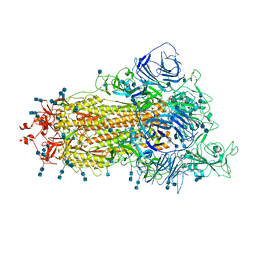 | | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351), uncleavable form, one RBD-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351), uncleavable form, one RBD-up conformation
To Be Published
|
|
7V7T
 
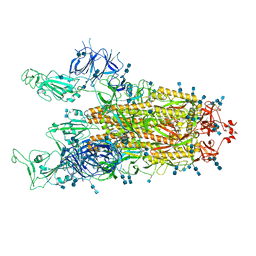 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), two RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), two RBD-up conformation 1
To Be Published
|
|
7V7Z
 
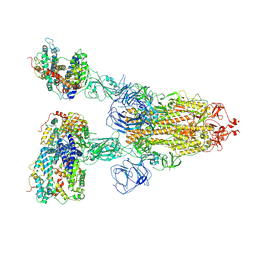 | | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form
To Be Published
|
|
7V86
 
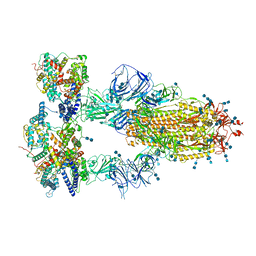 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form
To Be Published
|
|
7V89
 
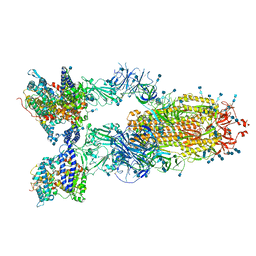 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 1
To Be Published
|
|
7V7V
 
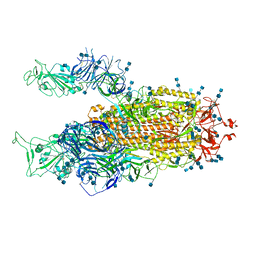 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), two RBD-up conformation 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), two RBD-up conformation 3
To Be Published
|
|
7V7G
 
 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), two RBD-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), two RBD-up conformation
To Be Published
|
|
7V8A
 
 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 2
To Be Published
|
|
7V8C
 
 | | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351), Cleavable form, one RBD-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351), Cleavable form, one RBD-up conformation
To Be Published
|
|
7V88
 
 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, two ACE2-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, two ACE2-bound form
To Be Published
|
|
7V7O
 
 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 1
To Be Published
|
|
7V7J
 
 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 3
To Be Published
|
|
7V7N
 
 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), all RBD-down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), all RBD-down conformation
To Be Published
|
|
7V78
 
 | | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), one RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), one RBD-up conformation 1
To Be Published
|
|
7V7A
 
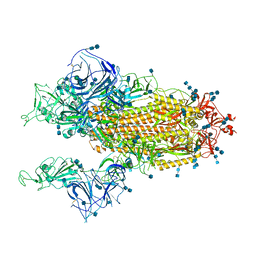 | | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), two RBD-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), two RBD-up conformation
To Be Published
|
|
7V7U
 
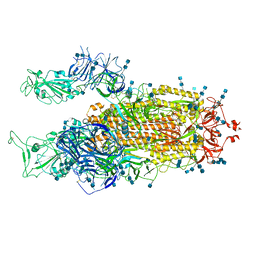 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), two RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), two RBD-up conformation 2
To Be Published
|
|
7V7R
 
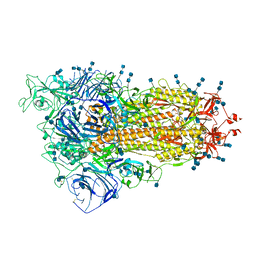 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 4
To Be Published
|
|
