3C8T
 
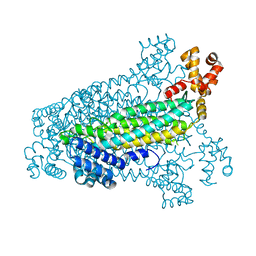 | | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1 | | 分子名称: | Fumarate lyase | | 著者 | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-02-13 | | 公開日 | 2008-03-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1.
To be Published
|
|
3C97
 
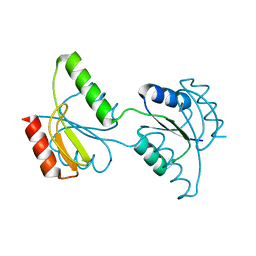 | | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae | | 分子名称: | Signal transduction histidine kinase | | 著者 | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-02-15 | | 公開日 | 2008-03-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae.
To be Published
|
|
3CYM
 
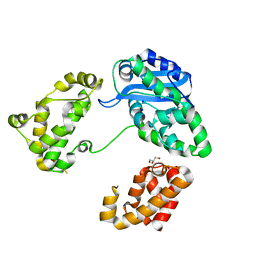 | | Crystal structure of protein BAD_0989 from Bifidobacterium adolescentis | | 分子名称: | GLYCEROL, SODIUM ION, Uncharacterized protein BAD_0989 | | 著者 | Patskovsky, Y, Ozyurt, S, Freeman, J, Chang, S, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-04-25 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of protein BAD_0989 from Bifidobacterium adolescentis.
To be Published
|
|
3CLW
 
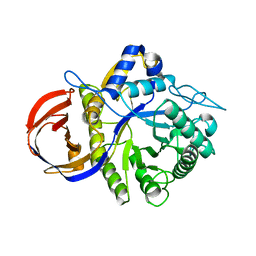 | | Crystal structure of conserved exported protein from Bacteroides fragilis | | 分子名称: | Conserved exported protein | | 著者 | Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Ozyurt, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-03-20 | | 公開日 | 2008-04-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | Crystal structure of conserved exported protein from Bacteroides fragilis.
To be Published
|
|
7D99
 
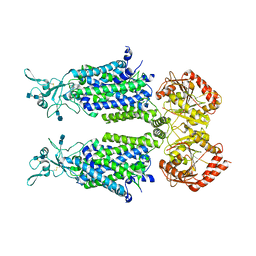 | | human potassium-chloride co-transporter KCC4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, POTASSIUM ION, ... | | 著者 | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | 登録日 | 2020-10-12 | | 公開日 | 2020-12-30 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
3FNR
 
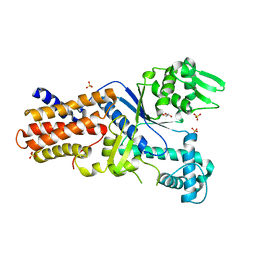 | | CRYSTAL STRUCTURE OF PUTATIVE ARGINYL T-RNA SYNTHETASE FROM Campylobacter jejuni; | | 分子名称: | Arginyl-tRNA synthetase, GLYCEROL, SULFATE ION | | 著者 | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-12-26 | | 公開日 | 2009-01-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF A PUTATIVE ARGINYL T-RNA SYNTHETASE FROM Campylobacter jejuni
To be Published
|
|
3GHY
 
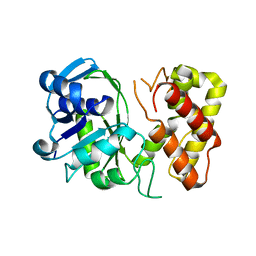 | | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2 | | 分子名称: | Ketopantoate reductase protein | | 著者 | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-03-04 | | 公開日 | 2009-03-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2
To be Published
|
|
3GFG
 
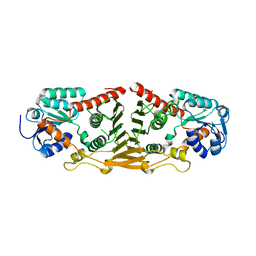 | | Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form | | 分子名称: | Uncharacterized oxidoreductase yvaA | | 著者 | Ramagopal, U.A, Toro, R, Gilmore, M, Chang, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-02-26 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Crystal structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form.
To be published
|
|
3DUP
 
 | | Crystal structure of mutt/nudix family hydrolase from rhodospirillum rubrum atcc 11170 | | 分子名称: | GLYCEROL, MutT/nudix family protein, PHOSPHATE ION | | 著者 | Patskovsky, Y, Ramagopal, U.A, Toro, R, Freeman, J, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-07-17 | | 公開日 | 2008-09-02 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Mutt/Nudix Family Hydrolase from Rhodospirillum Rubrum
To be Published
|
|
7D90
 
 | | human potassium-chloride co-transporter KCC3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 3 | | 著者 | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | 登録日 | 2020-10-12 | | 公開日 | 2020-12-30 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7D8Z
 
 | | human potassium-chloride co-transporter KCC2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 2 | | 著者 | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | 登録日 | 2020-10-11 | | 公開日 | 2020-12-30 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
3EZY
 
 | | Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Dehydrogenase | | 著者 | Ramagopal, U.A, Toro, R, Freeman, J, Chang, S, Maletic, M, Gheyi, T, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-10-24 | | 公開日 | 2009-01-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima
To be published
|
|
3GMF
 
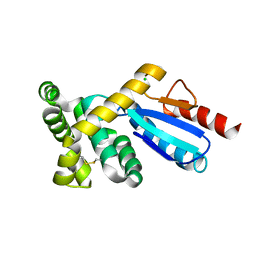 | | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans | | 分子名称: | CHLORIDE ION, Protein-disulfide isomerase | | 著者 | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-03-13 | | 公開日 | 2009-03-24 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans
To be Published
|
|
3GG9
 
 | | CRYSTAL STRUCTURE OF putative D-3-phosphoglycerate dehydrogenase oxidoreductase from Ralstonia solanacearum | | 分子名称: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | 著者 | Patskovsky, Y, Ramagopal, U, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-02-27 | | 公開日 | 2009-03-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Putative D-3-Phosphoglycerate Dehydrogenase from Ralstonia Solanacearum
To be Published
|
|
3GRZ
 
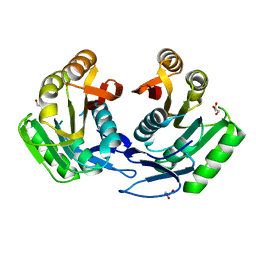 | | CRYSTAL STRUCTURE OF ribosomal protein L11 methylase FROM Lactobacillus delbrueckii subsp. bulgaricus | | 分子名称: | GLYCEROL, Ribosomal protein L11 methyltransferase | | 著者 | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-03-26 | | 公開日 | 2009-04-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN 11 METHYLASE FROM Lactobacillus delbrueckii subsp. bulgaricus
To be Published
|
|
3H7L
 
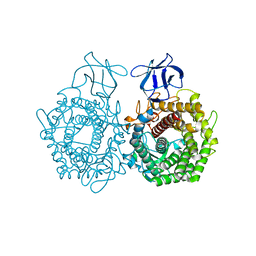 | | CRYSTAL STRUCTURE OF ENDOGLUCANASE-RELATED PROTEIN FROM Vibrio parahaemolyticus | | 分子名称: | ENDOGLUCANASE, GLYCEROL | | 著者 | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-04-27 | | 公開日 | 2009-05-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF ENDOGLUCANASE-RELATED PROTEIN FROM Vibrio parahaemolyticus
To be Published
|
|
3HCW
 
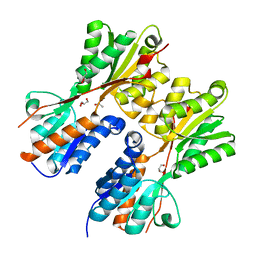 | | CRYSTAL STRUCTURE OF PROBABLE maltose operon transcriptional repressor malR FROM STAPHYLOCOCCUS AREUS | | 分子名称: | GLYCEROL, Maltose operon transcriptional repressor | | 著者 | Patskovsky, Y, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-05-06 | | 公開日 | 2009-05-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Maltose Operon Transcriptional Repressor from Staphylococcus Aureus
To be Published
|
|
3C19
 
 | | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19 | | 分子名称: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein MK0293 | | 著者 | Patskovsky, Y, Romero, R, Bonanno, J.B, Malashkevich, V, Dickey, M, Chang, S, Koss, J, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-01-22 | | 公開日 | 2008-02-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19.
To be Published
|
|
3C3M
 
 | | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1 | | 分子名称: | GLYCEROL, Response regulator receiver protein | | 著者 | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Dickey, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-01-28 | | 公開日 | 2008-02-05 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1.
To be Published
|
|
3CZ8
 
 | | Crystal structure of putative sporulation-specific glycosylase ydhD from Bacillus subtilis | | 分子名称: | GLYCEROL, Putative sporulation-specific glycosylase ydhD | | 著者 | Patskovsky, Y, Romero, R, Rutter, M, Chang, S, Maletic, M, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-04-28 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of putative glycosylase ydhD from Bacillus subtilis.
To be Published
|
|
6IRG
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class II | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (5.5 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRH
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class III | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
7JSN
 
 | | Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6 | | 分子名称: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, GUANOSINE-3',5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Gao, Y, Eskici, G, Ramachandran, S, Skiniotis, G, Cerione, R.A. | | 登録日 | 2020-08-15 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6.
Mol.Cell, 80, 2020
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
