2O19
 
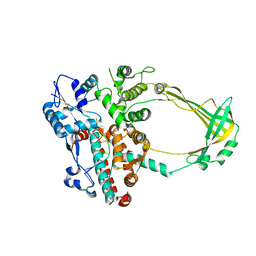 | | Structure of E. coli topoisomersae III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol at pH 5.5 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, DiGate, R, Mondragon, A. | | Deposit date: | 2006-11-28 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O59
 
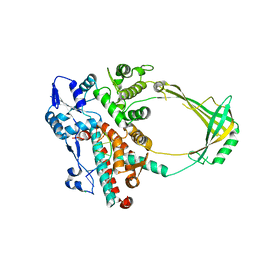 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol pH 8.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O5E
 
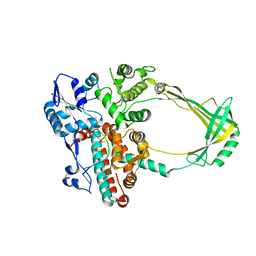 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glucose pH 7.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', CHLORIDE ION, DNA topoisomerase 3, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O54
 
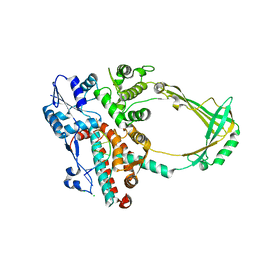 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol at pH 7.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, Digate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
4OQF
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIB-SR | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
2O5C
 
 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glucose pH 5.5 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', CHLORIDE ION, DNA topoisomerase 3, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
4PTL
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-GM | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-03-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
6XGU
 
 | | Crystal Structure of KRAS-Q61R (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF | | Descriptor: | GLYCEROL, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tran, T.H, Chan, A.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
7NHP
 
 | | Structure of PSII-I (PSII with Psb27, Psb28, and Psb34) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Zabret, J, Bohn, S, Schuller, S.K, Arnolds, O, Chan, A, Tajkhorshid, E, Stoll, R, Engel, B.D, Rudack, T, Schuller, J.M, Nowaczyk, M.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural insights into photosystem II assembly.
Nat.Plants, 7, 2021
|
|
7NHO
 
 | | Structure of PSII-M | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Zabret, J, Bohn, S, Schuller, S.K, Arnolds, O, Chan, A, Tajkhorshid, E, Stoll, R, Engel, B.D, Rudack, T, Schuller, J.M, Nowaczyk, M.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural insights into photosystem II assembly.
Nat.Plants, 7, 2021
|
|
7NHQ
 
 | | Structure of PSII-I prime (PSII with Psb28, and Psb34) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Zabret, J, Bohn, S, Schuller, S.K, Arnolds, O, Chan, A, Tajkhorshid, E, Stoll, R, Engel, B.D, Rudack, T, Schuller, J.M, Nowaczyk, M.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural insights into photosystem II assembly.
Nat.Plants, 7, 2021
|
|
3LZO
 
 | | Crystal Structure Analysis of the copper-reconstituted P19 protein from Campylobacter jejuni at 1.65 A at pH 10.0 | | Descriptor: | COPPER (II) ION, P19 protein, SULFATE ION | | Authors: | Doukov, T.I, Chan, A.C.K, Scofield, M, Ramin, A.B, Tom-Yew, S.A.L, Murphy, M.E.P. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Function of P19, a High-Affinity Iron Transporter of the Human Pathogen Campylobacter jejuni.
J.Mol.Biol., 401, 2010
|
|
3LZL
 
 | | Crystal Structure Analysis of the as-solated P19 protein from Campylobacter jejuni at 1.45 A at pH 9.0 | | Descriptor: | COPPER (II) ION, P19 protein, SULFATE ION | | Authors: | Doukov, T.I, Chan, A.C.K, Scofield, M, Ramin, A.B, Tom-Yew, S.A.L, Murphy, M.E.P. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and Function of P19, a High-Affinity Iron Transporter of the Human Pathogen Campylobacter jejuni.
J.Mol.Biol., 401, 2010
|
|
3LZN
 
 | | Crystal Structure Analysis of the apo P19 protein from Campylobacter jejuni at 1.59 A at pH 9 | | Descriptor: | P19 protein, SULFATE ION, ZINC ION | | Authors: | Doukov, T.I, Chan, A.C.K, Scofield, M, Ramin, A.B, Tom-Yew, S.A.L, Murphy, M.E.P. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and Function of P19, a High-Affinity Iron Transporter of the Human Pathogen Campylobacter jejuni.
J.Mol.Biol., 401, 2010
|
|
5A0E
 
 | | Crystal structure of cyclophilin D in complex with CsA analogue, JW47. | | Descriptor: | JW47, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE F, MITOCHONDRIAL | | Authors: | Warne, J, Pryce, G, Hill, J, Shi, X, Lenneras, F, Puentes, F, Kip, M, Hilditch, L, Walker, P, Simone, M, Chan, A.W.E, Towers, G, Coker, A.R, Duchen, M, Szabadkai, G, Baker, D, Selwood, D.L. | | Deposit date: | 2015-04-19 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Selective Inhibition of the Mitochondrial Permeability Transition Pore Protects Against Neuro-Degeneration in Experimental Multiple Sclerosis.
J.Biol.Chem., 291, 2016
|
|
6XJ6
 
 | |
6XM9
 
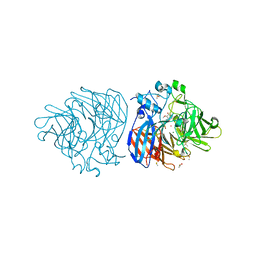 | | Crystal structure of vanillin bound to Co-LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETATE ION, COBALT (II) ION, ... | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6XJ7
 
 | |
6XM8
 
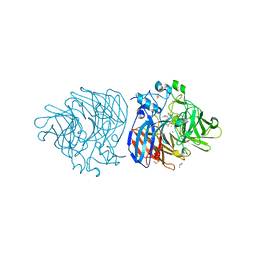 | | Crystal structure of lignostilbene bound to Co-LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | COBALT (II) ION, DIMETHYL SULFOXIDE, Dioxygenase, ... | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6XM7
 
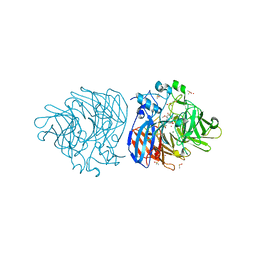 | | Crystal structure of DCA-S bound to Co-LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | (2E)-3-{4-hydroxy-3-[(E)-2-(4-hydroxy-3-methoxyphenyl)ethenyl]-5-methoxyphenyl}prop-2-enoic acid, COBALT (II) ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6XMA
 
 | | Crystal structure of iron-bound LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | Dioxygenase, FE (III) ION, SULFATE ION | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6XM6
 
 | | Crystal structure of cobalt-bound LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | COBALT (II) ION, Dioxygenase | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6E6I
 
 | | Crystal structure of 4-methyl HOPDA bound to LigY from Sphingobium sp. strain SYK-6 | | Descriptor: | (1Z,3E)-5-carboxy-3-methyl-5-oxo-1-phenylpenta-1,3-dien-1-olate, 2,2',3-trihydroxy-3'-methoxy-5,5'-dicarboxybiphenyl meta-cleavage compound hydrolase, ZINC ION | | Authors: | Kuatsjah, E, Chan, A.C, Hurst, T.E, Snieckus, V, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2018-07-24 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal- and Serine-Dependent Meta-Cleavage Product Hydrolases Utilize Similar Nucleophile-Activation Strategies
Acs Catalysis, 8, 2018
|
|
6OR7
 
 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with DNA AND (-)FTC-TP | | Descriptor: | DNA Primer 20-mer, DNA template 27-mer, MAGNESIUM ION, ... | | Authors: | Bertoletti, N, Chan, A.H, Anderson, K.S. | | Deposit date: | 2019-04-29 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: First crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine.
Protein Sci., 28, 2019
|
|
4WAA
 
 | | Crystal structure of Nix LIR-fused human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, Ravichandran, A.C, Dobson, R.C.J, Novak, I, Wakatsuki, S. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins.
Sci Rep, 7, 2017
|
|
