1S5P
 
 | | Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Eschericia coli. | | 分子名称: | HISTONE H4 (RESIDUES 12-19), NAD-dependent deacetylase, ZINC ION | | 著者 | Zhao, K, Chai, X, Marmorstein, R. | | 登録日 | 2004-01-21 | | 公開日 | 2004-03-23 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structure and Substrate Binding Properties of cobB, a Sir2 Homolog Protein Deacetylase from Eschericia coli.
J.Mol.Biol., 337, 2004
|
|
1SZD
 
 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | 登録日 | 2004-04-05 | | 公開日 | 2004-06-15 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1SZC
 
 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | 分子名称: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | 登録日 | 2004-04-05 | | 公開日 | 2004-06-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Q1A
 
 | | Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-acetyl ADP ribose and histone peptide | | 分子名称: | 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, HST2 protein, Histone H4, ... | | 著者 | Zhao, K, Chai, X, Marmorstein, R. | | 登録日 | 2003-07-18 | | 公開日 | 2003-11-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-Acetyl ADP ribose and histone peptide.
Structure, 11, 2003
|
|
1Q17
 
 | | Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-acetyl ADP ribose and histone peptide | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, HST2 protein, ... | | 著者 | Zhao, K, Chai, X, Marmorstein, R. | | 登録日 | 2003-07-18 | | 公開日 | 2003-11-18 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of the Yeast Hst2 Protein Deacetylase in Ternary Complex with 2'-O-Acetyl
ADP Ribose and Histone Peptide.
Structure, 11, 2003
|
|
1MX4
 
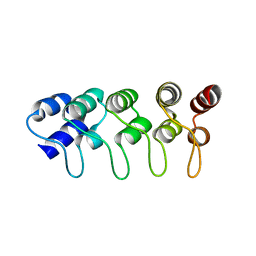 | | Structure of p18INK4c (F82Q) | | 分子名称: | Cyclin-dependent kinase 6 inhibitor | | 著者 | Marmorstein, R, Venkataramani, R.N, MacLachlan, T.K, Chai, X, El-Deiry, W.S. | | 登録日 | 2002-10-01 | | 公開日 | 2002-10-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based design of p18INK4c proteins with increased thermodynamic stability and cell cycle inhibitory activity
J.Biol.Chem., 277, 2002
|
|
1MX2
 
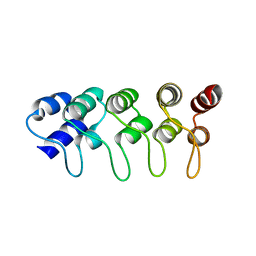 | | Structure of F71N mutant of p18INK4c | | 分子名称: | Cyclin-dependent kinase 6 inhibitor | | 著者 | Marmorstein, R, Venkataramani, R.N, MacLachlan, T.K, Chai, X, El-Deiery, W.S. | | 登録日 | 2002-10-01 | | 公開日 | 2002-10-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure-based design of p18INK4c proteins with increased thermodynamic stability and cell cycle inhibitory activity
J.Biol.Chem., 277, 2002
|
|
1MX6
 
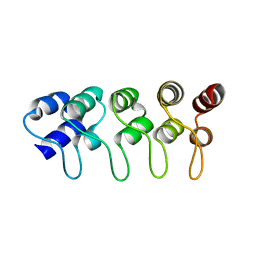 | | Structure of p18INK4c (F92N) | | 分子名称: | Cyclin-dependent kinase 6 inhibitor | | 著者 | Marmorstein, R, Venkataramani, R.N, MacLachlan, T.K, Chai, X, El-Deiry, W.S. | | 登録日 | 2002-10-01 | | 公開日 | 2002-10-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based design of p18INK4c proteins with increased thermodynamic stability and cell cycle inhibitory activity
J.Biol.Chem., 277, 2002
|
|
2IOO
 
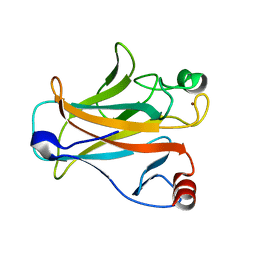 | | Crystal structure of the mouse p53 core domain | | 分子名称: | Cellular tumor antigen p53, ZINC ION | | 著者 | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | 登録日 | 2006-10-10 | | 公開日 | 2006-12-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2IOI
 
 | | Crystal structure of the mouse p53 core domain at 1.55 A | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular tumor antigen p53, ZINC ION | | 著者 | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | 登録日 | 2006-10-10 | | 公開日 | 2006-12-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2IOM
 
 | | Mouse p53 core domain soaked with 2-propanol | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular tumor antigen p53, ISOPROPYL ALCOHOL, ... | | 著者 | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | 登録日 | 2006-10-10 | | 公開日 | 2006-12-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
1HU8
 
 | | CRYSTAL STRUCTURE OF THE MOUSE P53 CORE DNA-BINDING DOMAIN AT 2.7A RESOLUTION | | 分子名称: | CELLULAR TUMOR ANTIGEN P53, ZINC ION | | 著者 | Zhao, K, Chai, X, Johnston, K, Clements, A, Marmorstein, R. | | 登録日 | 2001-01-04 | | 公開日 | 2001-07-04 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of the mouse p53 core DNA-binding domain at 2.7 A resolution.
J.Biol.Chem., 276, 2001
|
|
1NFJ
 
 | | Structure of a Sir2 substrate, alba, reveals a mechanism for deactylation-induced enhancement of DNA-binding | | 分子名称: | conserved hypothetical protein AF1956 | | 著者 | Zhao, K, Chai, X, Marmorstein, R. | | 登録日 | 2002-12-15 | | 公開日 | 2003-08-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a Sir2 substrate, alba, reveals a mechanism for deacetylation-induced enhancement of DNA-binding
J.Biol.Chem., 278, 2003
|
|
1Q14
 
 | | Structure and autoregulation of the yeast Hst2 homolog of Sir2 | | 分子名称: | CHLORIDE ION, HST2 protein, ZINC ION | | 著者 | Zhao, K, Chai, X, Clements, A, Marmorstein, R. | | 登録日 | 2003-07-18 | | 公開日 | 2003-09-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and autoregulation of the Yeast Hst2 homolog of Sir2
Nat.Struct.Biol., 10, 2003
|
|
1NFH
 
 | | Structure of a Sir2 substrate, alba, reveals a mechanism for deactylation-induced enhancement of DNA-binding | | 分子名称: | conserved hypothetical protein AF1956 | | 著者 | Zhao, K, Chai, X, Marmorstein, R. | | 登録日 | 2002-12-15 | | 公開日 | 2003-08-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure of a Sir2 substrate, Alba, reveals a mechanism for deacetylation-induced enhancement of DNA-binding
J.Biol.Chem., 278, 2003
|
|
