3EDV
 
 | |
1N11
 
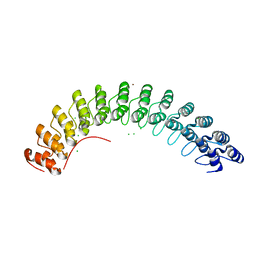 | | D34 REGION OF HUMAN ANKYRIN-R AND LINKER | | Descriptor: | Ankyrin, BROMIDE ION, CHLORIDE ION | | Authors: | Michaely, P, Tomchick, D.R, Machius, M, Anderson, R.G.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a 12 ANK repeat stack from human ankyrinR
Embo J., 21, 2002
|
|
3PDS
 
 | | Irreversible Agonist-Beta2 Adrenoceptor Complex | | Descriptor: | 8-hydroxy-5-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(3-sulfanylpropoxy)phenyl]ethyl}amino)ethyl]quinolin-2(1H)-one, CHOLESTEROL, Fusion protein Beta-2 adrenergic receptor/Lysozyme, ... | | Authors: | Rosenbaum, D.M, Zhang, C, Lyons, J.A, Holl, R, Aragao, D, Arlow, D.H, Rasmussen, S.G.F, Choi, H.-J, DeVree, B.T, Sunahara, R.K, Chae, P.S, Gellman, S.H, Dror, R.O, Shaw, D.E, Weis, W.I, Caffrey, M, Gmeiner, P, Kobilka, B.K. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and function of an irreversible agonist-beta(2) adrenoceptor complex
Nature, 469, 2011
|
|
3P0G
 
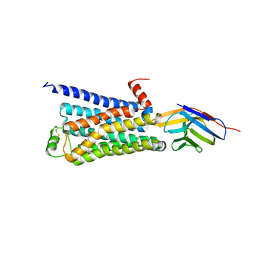 | | Structure of a nanobody-stabilized active state of the beta2 adrenoceptor | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Beta-2 adrenergic receptor, Lysozyme, ... | | Authors: | Rasmussen, S.G.F, Choi, H.-J, Fung, J.J, Pardon, E, Casarosa, P, Chae, P.S, DeVree, B.T, Rosenbaum, D.M, Thian, F.S, Kobilka, T.S, Schnapp, A, Konetzki, I, Sunahara, R.K, Gellman, S.H, Pautsch, A, Steyaert, J, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2010-09-28 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a nanobody-stabilized active state of the b2 adrenoceptor
Nature, 469, 2011
|
|
4DAJ
 
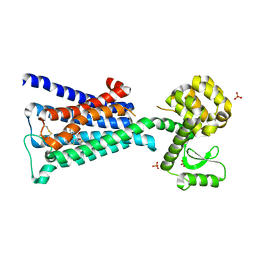 | | Structure of the M3 Muscarinic Acetylcholine Receptor | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3, Lysozyme, ... | | Authors: | Kruse, A.C, Hu, J, Pan, A.C, Arlow, D.H, Rosenbaum, D.M, Rosemond, E, Green, H.F, Liu, T, Chae, P.S, Dror, R.O, Shaw, D.E, Weis, W.I, Wess, J, Kobilka, B. | | Deposit date: | 2012-01-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and dynamics of the M3 muscarinic acetylcholine receptor.
Nature, 482, 2012
|
|
3SN6
 
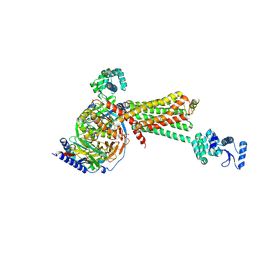 | | Crystal structure of the beta2 adrenergic receptor-Gs protein complex | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid antibody VHH fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Rasmussen, S.G.F, DeVree, B.T, Zou, Y, Kruse, A.C, Chung, K.Y, Kobilka, T.S, Thian, F.S, Chae, P.S, Pardon, E, Calinski, D, Mathiesen, J.M, Shah, S.T.A, Lyons, J.A, Caffrey, M, Gellman, S.H, Steyaert, J, Skiniotis, G, Weis, W.I, Sunahara, R.K, Kobilka, B.K. | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the beta2 adrenergic receptor-Gs protein complex
Nature, 477, 2011
|
|
3ZX3
 
 | | Crystal Structure and Domain Rotation of NTPDase1 CD39 | | Descriptor: | ACETIC ACID, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 1, ... | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Evidence for a Domain Motion in Rat Nucleoside Triphosphate Diphosphohydrolase (Ntpdase) 1.
J.Mol.Biol., 415, 2012
|
|
3ZX0
 
 | | NTPDase1 in complex with Heptamolybdate | | Descriptor: | ACETIC ACID, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 1, ... | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic evidence for a domain motion in rat nucleoside triphosphate diphosphohydrolase (NTPDase) 1.
J. Mol. Biol., 415, 2012
|
|
3ZX2
 
 | | NTPDase1 in complex with Decavanadate | | Descriptor: | ACETIC ACID, CHLORIDE ION, DECAVANADATE, ... | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic Evidence for a Domain Motion in Rat Nucleoside Triphosphate Diphosphohydrolase (Ntpdase) 1.
J.Mol.Biol., 415, 2012
|
|
4BRH
 
 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with MG AND THIAMINE PHOSPHOVANADATE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DECAVANADATE, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystallographic snapshots along the reaction pathway of nucleoside triphosphate diphosphohydrolases.
Structure, 21, 2013
|
|
4BRE
 
 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with transition state mimic adenosine 5'phosphovanadate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-PHOSPHOVANADATE, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRL
 
 | | Legionella pneumophila NTPDase1 crystal form III (closed) in complex with transition state mimic guanosine 5'-phosphovanadate | | Descriptor: | CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, GUANOSINE-5'-PHOSPHOVANADATE, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BR9
 
 | | Legionella pneumophila NTPDase1 crystal form II, closed, apo | | Descriptor: | ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, GLYCEROL, SULFATE ION | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRF
 
 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with a distorted orthomolybdate ion and AMP | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRG
 
 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with MG GMPPNP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRQ
 
 | | LEGIONELLA PNEUMOPHILA NTPDASE1 CRYSTAL FORM II, CLOSED, IN COMPLEX WITH TWO PHOSPHATES BOUND TO ACTIVE SITE MG AND PRODUCT AMP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRC
 
 | | Legionella pneumophila NTPDase1 crystal form II, closed, Mg AMPNP complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-O-[(R)-hydroxy(phosphonoamino)phosphoryl]adenosine, CHLORIDE ION, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRK
 
 | | Legionella pneumophila NTPDase1 N302Y variant crystal form III (closed) in complex with MG UMPPNP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]uridine, CHLORIDE ION, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BR4
 
 | | Legionella pneumophila NTPDase1 crystal form I, open, apo | | Descriptor: | CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, MAGNESIUM ION | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-03 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRM
 
 | | Sulfur SAD phasing of the Legionella pneumophila NTPDase1 - crystal form III (closed) in complex with sulfate | | Descriptor: | CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, SULFATE ION | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BR0
 
 | | rat NTPDase2 in complex with Ca AMPNP | | Descriptor: | 5'-O-[(R)-hydroxy(phosphonoamino)phosphoryl]adenosine, CALCIUM ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-03 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRA
 
 | | Legionella pneumophila NTPDase1 crystal form II, closed, Mg AMPPNP complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, GLYCEROL, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BTT
 
 | | factor Xa in complex with the dual thrombin-FXa inhibitor 31. | | Descriptor: | CALCIUM ION, COAGULATION FACTOR X, COAGULATION FACTOR X LIGHT CHAIN, ... | | Authors: | Meneyrol, J, Follmann, M, Lassalle, G, Wehner, V, Barre, G, Rousseaux, T, Altenburger, J.M, Petit, F, Bocskei, Z, Stehlin-Gaon, C, Schreuder, H, Alet, N, Herault, J.-P, Millet, L, Dol, F, Hasbrand, C, Schaeffer, P, Sadoun, F, Klieber, S, Briot, C, Bono, F, Herbert, J.-M. | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | 5-Chlorothiophene-2-Carboxylic Acid [(S)-2-[2-Methyl-3-(2-Oxopyrrolidin-1-Yl)Benzenesulfonylamino]-3-(4-Methylpiperazin-1-Yl)-3-Oxopropyl]Amide (Sar107375), a Selective and Potent Orally Active Dual Thrombin and Factor Xa Inhibitor.
J.Med.Chem., 56, 2013
|
|
4BTI
 
 | | factor Xa in complex with the dual thrombin-FXa inhibitor 58. | | Descriptor: | 5-Chloro-thiophene-2-carboxylic acid [(S)-2-[2-difluoromethoxy-3-(2-oxo-piperidin-1-yl)-benzenesulfonylamino]-3-((S)-3-dimethylamino-pyrrolidin-1-yl)-3-oxo-propyl]-amide, CALCIUM ION, COAGULATION FACTOR X HEAVY CHAIN, ... | | Authors: | Meneyrol, J, Follmann, M, Lassalle, G, Wehner, V, Barre, G, Rousseaux, T, Altenburger, J.M, Petit, F, Bocskei, Z, Stehlin-Gaon, C, Schreuder, H, Alet, N, Herault, J.-P, Millet, L, Dol, F, Hasbrand, C, Schaeffer, P, Sadoun, F, Klieber, S, Briot, C, Bono, F, Herbert, J.-M. | | Deposit date: | 2013-06-18 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 5-Chlorothiophene-2-Carboxylic Acid [(S)-2-[2-Methyl-3-(2-Oxopyrrolidin-1-Yl)Benzenesulfonylamino]-3-(4-Methylpiperazin-1-Yl)-3-Oxopropyl]Amide (Sar107375), a Selective and Potent Orally Active Dual Thrombin and Factor Xa Inhibitor.
J.Med.Chem., 56, 2013
|
|
4BR2
 
 | | rat NTPDase2 in complex with Ca UMPPNP | | Descriptor: | 5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]uridine, CALCIUM ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-03 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
