7B0X
 
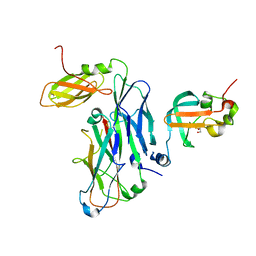 | | Crystal structure of the ternary complex of the E. coli type 1 pilus proteins FimC, FimI and the N-terminal domain of FimD | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein FimC, Fimbrin-like protein FimI, ... | | Authors: | Scharer, M.A, Zigova, Z, Giese, C, Puorger, C, Ignatov, O, Capitani, G, Glockshuber, R. | | Deposit date: | 2020-11-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
8ATL
 
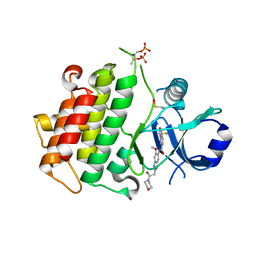 | | Discovery of IRAK4 Inhibitor 23 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-[6-methoxy-2-(2-morpholin-4-yl-2-oxidanylidene-ethyl)indazol-5-yl]-6-[(1~{R})-2,2,2-tris(fluoranyl)-1-oxidanyl-ethyl]pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-08-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.464 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
8ATN
 
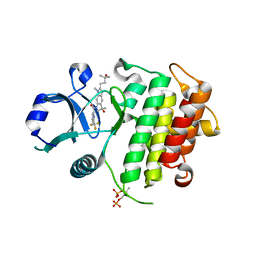 | | Discovery of IRAK4 Inhibitor 38 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-[3-methyl-2-(3-methyl-3-oxidanyl-butyl)-6-(2-oxidanylpropan-2-yl)benzimidazol-5-yl]-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-08-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
6ZUG
 
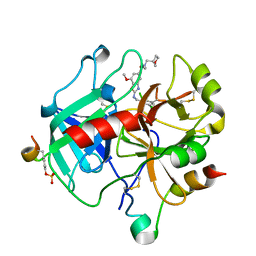 | | Crystal Structure of Thrombin in complex with compound10 | | Descriptor: | 2-[(3-chlorophenyl)methylamino]-7-methoxy-~{N}-[[(3~{S})-oxolan-3-yl]methyl]-~{N}-propyl-1,3-benzoxazole-5-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-22 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
6ZUU
 
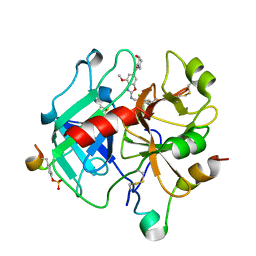 | | Crystal structure of Thrombin in complex with compound30 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, Prothrombin, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
6ZV7
 
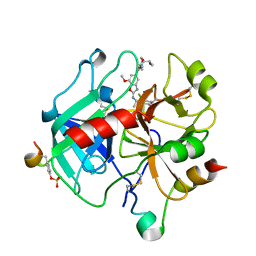 | | Crystal Structure of Thrombin in complex with compound42b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, Prothrombin, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
6ZV8
 
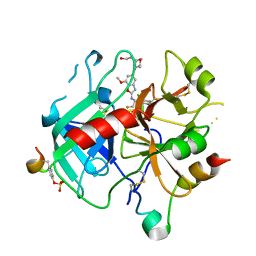 | | Crystal Structure of Thrombin in complex with compound51 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, Prothrombin, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
8ATB
 
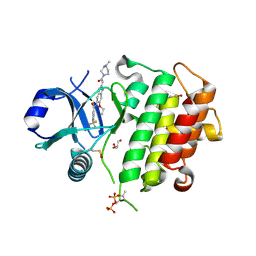 | | Discovery of IRAK4 Inhibitor 16 | | Descriptor: | GLYCEROL, Interleukin-1 receptor-associated kinase 4, ~{N}-[6-ethoxy-2-[2-(4-methylpiperazin-1-yl)-2-oxidanylidene-ethyl]indazol-5-yl]-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-08-22 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
6ZUN
 
 | | Crystal Structure of Thrombin in complex with compound20a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, Prothrombin, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
6ZUX
 
 | | Crystal Structure of Thrombin in complex with compound42a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin-2, Prothrombin, ... | | Authors: | Schafer, M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
J.Med.Chem., 63, 2020
|
|
7Z90
 
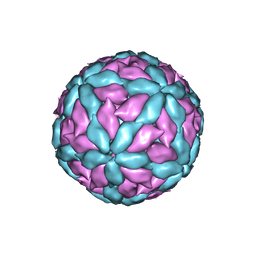 | | Leishmania RNA virus 1 virion | | Descriptor: | Capsid protein,Major capsid protein | | Authors: | Prochazkova, M, Plevka, P. | | Deposit date: | 2022-03-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Virion structure of Leishmania RNA virus 1.
Virology, 577, 2022
|
|
4XZQ
 
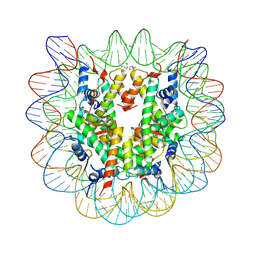 | | Nucleosome disassembly by RSC and SWI/SNF is enhanced by H3 acetylation near the nucleosome dyad axis | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Dechassa, M.L, Luger, K, Chatterjee, N, North, J.A, Manohar, M, Prasad, R, Ottessen, J.J, Poirier, M.G, Bartholomew, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Histone Acetylation near the Nucleosome Dyad Axis Enhances Nucleosome Disassembly by RSC and SWI/SNF.
Mol.Cell.Biol., 35, 2015
|
|
6USO
 
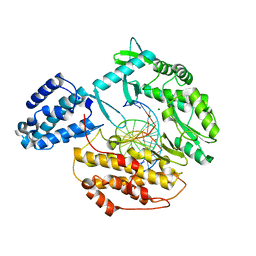 | |
6USP
 
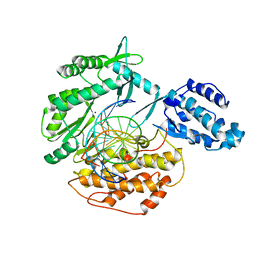 | | Telomerase Reverse Transcriptase product complex, TERT:DNA | | Descriptor: | DNA (5'-D(P*GP*GP*TP*CP*AP*GP*GP*TP*CP*AP*GP*GP*TP*CP*AP*G)-3'), DNA/RNA (5'-R(*CP*UP*GP*AP*CP*CP*UP*GP*AP*C)-D(P*CP*T)-R(P*GP*AP*C)-D(P*C)-3'), MAGNESIUM ION, ... | | Authors: | Schaich, M.A, Freudenthal, B.D. | | Deposit date: | 2019-10-28 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Mechanisms of nucleotide selection by telomerase.
Elife, 9, 2020
|
|
6USQ
 
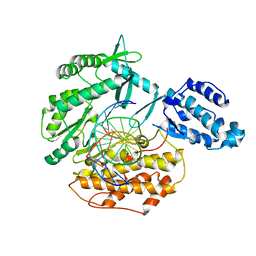 | | Telomerase Reverse Transcriptase binary complex with Y256A mutation, TERT:DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*AP*GP*GP*TP*CP*AP*GP*GP*TP*CP*A)-3'), DNA/RNA (5'-R(*CP*UP*GP*AP*CP*CP*UP*GP*AP*C)-D(P*CP*T)-R(P*GP*AP*C)-D(P*C)-3'), MAGNESIUM ION, ... | | Authors: | Schaich, M.A, Freudenthal, B.D, Khoang, T.H. | | Deposit date: | 2019-10-28 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Mechanisms of nucleotide selection by telomerase.
Elife, 9, 2020
|
|
4Z66
 
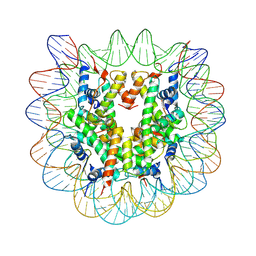 | | Nucleosome disassembly by RSC and SWI/SNF is enhanced by H3 acetylation near the nucleosome dyad axis | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Dechassa, M.L, Luger, K, Chatterjee, N, North, J.A, Manohar, M, Prasad, R, Ottessen, J.J, Poirier, M.G, Bartholomew, B. | | Deposit date: | 2015-04-03 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Histone Acetylation near the Nucleosome Dyad Axis Enhances Nucleosome Disassembly by RSC and SWI/SNF.
Mol.Cell.Biol., 35, 2015
|
|
6USR
 
 | |
6XJ1
 
 | |
7MZV
 
 | | Structure of yeast pseudouridine synthase 7 (PUS7) | | Descriptor: | Multisubstrate pseudouridine synthase 7, SULFATE ION | | Authors: | Purchal, M, Koutmos, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Pseudouridine synthase 7 is an opportunistic enzyme that binds and modifies substrates with diverse sequences and structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4YS3
 
 | | Nucleosome disassembly by RSC and SWI/SNF is enhanced by H3 acetylation near the nucleosome dyad axis | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Dechassa, M.L, Luger, K, Chatterjee, N, North, J.A, Manohar, M, Prasad, R, Ottessen, J.J, Poirier, M.G, Bartholomew, B. | | Deposit date: | 2015-03-16 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Histone Acetylation near the Nucleosome Dyad Axis Enhances Nucleosome Disassembly by RSC and SWI/SNF.
Mol.Cell.Biol., 35, 2015
|
|
4TXO
 
 | | Crystal structure of the mixed disulfide complex of thioredoxin-like TlpAs(C110S) and copper chaperone ScoIs(C74S) | | Descriptor: | Blr1131 protein, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Scharer, M.A, Abicht, H.K, Glockshuber, R, Hennecke, H. | | Deposit date: | 2014-07-04 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How Periplasmic Thioredoxin TlpA Reduces Bacterial Copper Chaperone ScoI and Cytochrome Oxidase Subunit II (CoxB) Prior to Metallation.
J.Biol.Chem., 289, 2014
|
|
4U47
 
 | | Octameric RNA duplex soaked in terbium(III)chloride | | Descriptor: | RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3'), TERBIUM(III) ION | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-23 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
4U3P
 
 | | Octameric RNA duplex co-crystallized with strontium(II)chloride | | Descriptor: | RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3'), STRONTIUM ION | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
4U3R
 
 | | Octameric RNA duplex co-crystallized with cobalt(II)chloride | | Descriptor: | COBALT (II) ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
8B3W
 
 | | Structure of metacyclic VSG (mVSG) 1954 from Trypanosoma brucei | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, Variant surface glycoprotein 1954 | | Authors: | Chandra, M, Stebbins, C.E. | | Deposit date: | 2022-09-17 | | Release date: | 2023-01-25 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (1.681 Å) | | Cite: | Structural similarities between the metacyclic and bloodstream form variant surface glycoproteins of the African trypanosome.
Plos Negl Trop Dis, 17, 2023
|
|
