1PV3
 
 | | NMR Solution Structure of the Avian FAT-domain of Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Prutzman, K.C, Gao, G, King, M.L, Iyer, V.V, Mueller, G.A, Schaller, M.D, Campbell, S.L. | | Deposit date: | 2003-06-26 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Focal Adhesion Targeting Domain of Focal Adhesion Kinase Contains a Hinge Region that Modulates Tyrosine 926 Phosphorylation.
STRUCTURE, 12, 2004
|
|
1Q5V
 
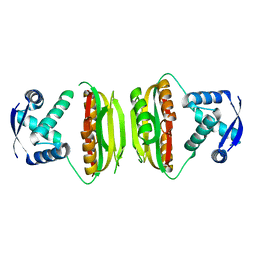 | | Apo-NikR | | Descriptor: | Nickel responsive regulator | | Authors: | Schreiter, E.R, Sintchak, M.D, Guo, Y, Chivers, P.T, Sauer, R.T, Drennan, C.L. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Nickel-Responsive Transcription Factor NikR
Nat.Struct.Biol., 10, 2003
|
|
1Q5Y
 
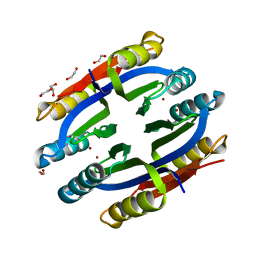 | | Nickel-Bound C-terminal Regulatory Domain of NikR | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Nickel responsive regulator | | Authors: | Schreiter, E.R, Sintchak, M.D, Guo, Y, Chivers, P.T, Sauer, R.T, Drennan, C.L. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Nickel-Responsive Transcription Factor NikR
Nat.Struct.Biol., 10, 2003
|
|
1BSK
 
 | | ZINC DEFORMYLASE INHIBITOR COMPLEX FROM E.COLI | | Descriptor: | (S)-2-(PHOSPHONOXY)CAPROYL-L-LEUCYL-P-NITROANILIDE, PHOSPHATE ION, PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Hao, B, Gong, W, Rajagopalan, P.T, Hu, Y, Pei, D, Chan, M.K. | | Deposit date: | 1998-08-28 | | Release date: | 2000-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the design of antibiotics targeting peptide deformylase.
Biochemistry, 38, 1999
|
|
4CST
 
 | | Crystal structure of FimH in complex with 3'-Chloro-4'-(alpha-D-mannopyranosyloxy)-biphenyl-4-carbonitrile | | Descriptor: | 3'-chloro-4'-(alpha-D-mannopyranosyloxy)biphenyl-4-carbonitrile, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
1JQK
 
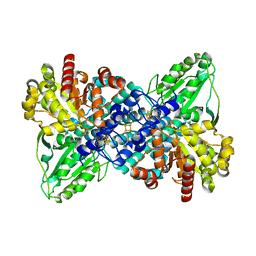 | | Crystal structure of carbon monoxide dehydrogenase from Rhodospirillum rubrum | | Descriptor: | FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Drennan, C.L, Heo, J, Sintchak, M.D, Schreiter, E, Ludden, P.W. | | Deposit date: | 2001-08-07 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Life on carbon monoxide: X-ray structure of Rhodospirillum rubrum Ni-Fe-S carbon monoxide dehydrogenase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2A6O
 
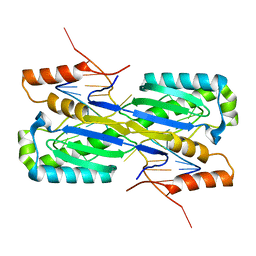 | | Crystal Structure of the ISHp608 Transposase in Complex with Stem-loop DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*TP*AP*GP*CP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*GP*A)-3', ISHp608 Transposase | | Authors: | Ronning, D.R, Guynet, C, Ton-Hoang, B, Perez, Z.N, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active site sharing and subterminal hairpin recognition in a new class of DNA transposases.
Mol.Cell, 20, 2005
|
|
2W26
 
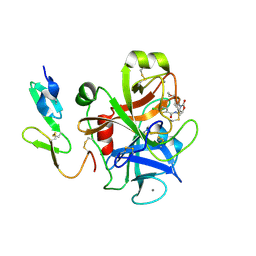 | | Factor Xa in complex with BAY59-7939 | | Descriptor: | 5-chloro-N-({(5S)-2-oxo-3-[4-(3-oxomorpholin-4-yl)phenyl]-1,3-oxazolidin-5-yl}methyl)thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION | | Authors: | Roehrig, S, Straub, A, Pohlmann, J, Lampe, T, Pernerstorfer, J, Schlemmer, K, Reinemer, P, Perzborn, E, Schaefer, M. | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of the Novel Antithrombotic Agent 5-Chloro-N-({(5S)-2-Oxo-3- [4-(3-Oxomorpholin-4-Yl)Phenyl]-1,3-Oxazolidin-5-Yl}Methyl)Thiophene-2- Carboxamide (Bay 59-7939): An Oral, Direct Factor Xa Inhibitor.
J.Med.Chem., 48, 2005
|
|
4CZ4
 
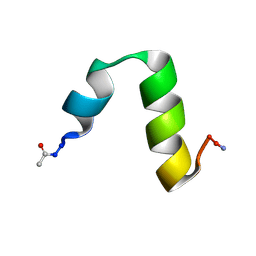 | | HP24stab derived from the villin headpiece subdomain | | Descriptor: | VILLIN-1 | | Authors: | Hocking, H, Haese, F, Madl, T, Zacharias, M, Rief, M, Zoldak, G. | | Deposit date: | 2014-04-16 | | Release date: | 2015-02-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Compact Native 24-Residue Supersecondary Structure Derived from the Villin Headpiece Subdomain.
Biophys.J., 108, 2015
|
|
4CZ3
 
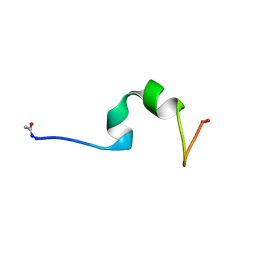 | | HP24wt derived from the villin headpiece subdomain | | Descriptor: | VILLIN-1 | | Authors: | Hocking, H, Haese, F, Madl, T, Zacharias, M, Rief, M, Zoldak, G. | | Deposit date: | 2014-04-16 | | Release date: | 2015-02-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Compact Native 24-Residue Supersecondary Structure Derived from the Villin Headpiece Subdomain.
Biophys.J., 108, 2015
|
|
2A6M
 
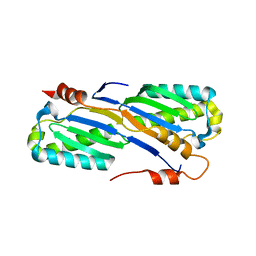 | | Crystal Structure of the ISHp608 Transposase | | Descriptor: | ISHp608 transposase | | Authors: | Ronning, D.R, Guynet, C, Ton-Hoang, B, Perez, Z.N, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active site sharing and subterminal hairpin recognition in a new class of DNA transposases.
Mol.Cell, 20, 2005
|
|
1TCO
 
 | | TERNARY COMPLEX OF A CALCINEURIN A FRAGMENT, CALCINEURIN B, FKBP12 AND THE IMMUNOSUPPRESSANT DRUG FK506 (TACROLIMUS) | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, FE (III) ION, ... | | Authors: | Griffith, J.P, Kim, J.L, Kim, E.E, Sintchak, M.D, Thomson, J.A, Fitzgibbon, M.J, Fleming, M.A, Caron, P.R, Hsiao, K, Navia, M.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex.
Cell(Cambridge,Mass.), 82, 1995
|
|
1SXD
 
 | | Solution Structure of the Pointed (PNT) Domain from mGABPa | | Descriptor: | GA repeat binding protein, alpha | | Authors: | Mackereth, C.D, Schaerpf, M, Gentile, L.N, MacIntosh, S.E, Slupsky, C.M, McIntosh, L.P. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Diversity in Structure and Function of the Ets Family PNT Domains.
J.Mol.Biol., 342, 2004
|
|
1JJH
 
 | |
1T3P
 
 | | Half-sandwich arene ruthenium(II)-enzyme complex | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | McNae, I.W, Fishburne, K, Habtemariam, A, Hunter, T.M, Melchart, M, Wang, F, Walkinshaw, M.D, Sadler, P.J. | | Deposit date: | 2004-04-27 | | Release date: | 2005-07-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Half-sandwich arene ruthenium(II)-enzyme complex
CHEM.COMMUN.(CAMB.), 16, 2004
|
|
1T6F
 
 | | Crystal Structure of the Coiled-coil Dimerization Motif of Geminin | | Descriptor: | Geminin | | Authors: | Thepaut, M, Maiorano, D, Guichou, J.-F, Auge, M.-T, Dumas, C, Mechali, M, Padilla, A. | | Deposit date: | 2004-05-06 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of the Coiled-coil Dimerization Motif of Geminin: Structural and Functional Insights on DNA Replication Regulation
J.Mol.Biol., 342, 2004
|
|
2LDD
 
 | | Solution structure of the estrogen receptor-binding stapled peptide SP6 (Ac-EKHKILXRLLXDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP6 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2J0M
 
 | | Crystal structure a two-chain complex between the FERM and kinase domains of focal adhesion kinase. | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, FOCAL ADHESION KINASE 1 | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase.
Cell(Cambridge,Mass.), 129, 2007
|
|
2J0J
 
 | | Crystal structure of a fragment of focal adhesion kinase containing the FERM and kinase domains. | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, FOCAL ADHESION KINASE 1 | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase
Cell(Cambridge,Mass.), 129, 2007
|
|
2J0K
 
 | | Crystal structure of a fragment of focal adhesion kinase containing the FERM and kinase domains. | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, FOCAL ADHESION KINASE 1 | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase.
Cell(Cambridge,Mass.), 129, 2007
|
|
2J0L
 
 | | Crystal structure of a the active conformation of the kinase domain of focal adhesion kinase with a phosphorylated activation loop. | | Descriptor: | FOCAL ADHESION KINASE 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase
Cell(Cambridge,Mass.), 129, 2007
|
|
2JV3
 
 | |
2K28
 
 | | Solution NMR structure of the chromo domain of the chromobox protein homolog 4 | | Descriptor: | E3 SUMO-protein ligase CBX4 | | Authors: | Kaustov, L, Lemak, A, Quyang, H, Fares, C, Gutmanas, A, Ravichandran, M, Loppnau, P, Bountra, C, Weigelt, J, Edwards, A.M, Min, J, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the chromo domain of the chromobox protein homolog 4.
To be Published
|
|
2JUO
 
 | | GABPa OST domain | | Descriptor: | GA-binding protein alpha chain | | Authors: | Kang, H, Nelson, M.L, Mackereth, C.D, Schaerpf, M, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2007-08-31 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Identification and structural characterization of a CBP/p300-binding domain from the ETS family transcription factor GABP alpha
J.Mol.Biol., 377, 2008
|
|
2K1B
 
 | | Solution NMR structure of the chromo domain of the chromobox protein homolog 7 | | Descriptor: | Chromobox protein homolog 7 | | Authors: | Kaustov, L, Lemak, A, Quyang, H, Gutmanas, A, Fares, C, Bountra, C, Weigelt, J, Loppnau, P, Ravichandran, M, Edwards, A.M, Min, J, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the chromobox protein homolog 7.
To be Published
|
|
