2WT8
 
 | |
3ZN2
 
 | | protein engineering of halohydrin dehalogenase | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ACETATE ION, HALOHYDRIN DEHALOGENASE, ... | | Authors: | Schallmey, M, Jekel, P, Tang, L, Majeric-Elenkov, M, Hoeffken, H.W, Hauer, B, Janssen, D.B. | | Deposit date: | 2013-02-13 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Single Point Mutation Enhances Hydroxynitrile Synthesis by Halohydrin Dehalogenase.
Enzyme.Microb.Technol., 70, 2015
|
|
2LD0
 
 | |
2J9M
 
 | | Crystal Structure of CDK2 in complex with Macrocyclic Aminopyrimidine | | Descriptor: | 6-BROMO-13-THIA-2,4,8,12,19-PENTAAZATRICYCLO[12.3.1.1~3,7~]NONADECA-1(18),3(19),4,6,14,16-HEXAENE 13,13-DIOXIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Schaefer, M, Luecking, U, Siemeister, G, Briem, H, Krueger, M, Lienau, P, Jautelat, R. | | Deposit date: | 2006-11-13 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Macrocyclic Aminopyrimidines as Multitarget Cdk and Vegf-R Inhibitors with Potent Antiproliferative Activities.
Chemmedchem, 2, 2007
|
|
2LD2
 
 | |
2LRE
 
 | | The solution structure of the dimeric Acanthaporin | | Descriptor: | Acanthaporin | | Authors: | Michalek, M, Soennichsen, F.D, Wechselberger, R, Dingley, A.J, Wienk, H, Simanski, M, Herbst, R, Lorenzen, I, Marciano-Cabral, F, Gelhaus, C, Groetzinger, J, Leippe, M. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba.
Nat.Chem.Biol., 9, 2013
|
|
2LRD
 
 | | The solution structure of the monomeric Acanthaporin | | Descriptor: | Acanthaporin | | Authors: | Michalek, M, Soennichsen, F.D, Wechselberger, R, Dingley, A.J, Wienk, H, Simanski, M, Herbst, R, Lorenzen, I, Marciano-Cabral, F, Gelhaus, C, Groetzinger, J, Leippe, M. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba.
Nat.Chem.Biol., 9, 2013
|
|
4M1E
 
 | | Crystal structure of purine nucleoside phosphorylase I from Planctomyces limnophilus DSM 3776, NYSGRC Target 029364. | | Descriptor: | ADENINE, PYRIDINE-2-CARBOXYLIC ACID, Purine nucleoside phosphorylase, ... | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase I from Planctomyces limnophilus DSM 3776, NYSGRC Target 029364.
To be Published
|
|
4MCH
 
 | | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with 6-hydroxy-1-naphthoic acid, NYSGRC Target 029520. | | Descriptor: | 6-hydroxynaphthalene-1-carboxylic acid, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with 6-hydroxy-1-naphthoic acid, NYSGRC Target 029520.
To be Published
|
|
4M0K
 
 | | Crystal structure of adenine phosphoribosyltransferase from Rhodothermus marinus DSM 4252, NYSGRC Target 029775. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, CALCIUM ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of adenine phosphoribosyltransferase from Rhodothermus marinus DSM 4252, NYSGRC Target 029775.
To be Published
|
|
4MAR
 
 | | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279 complexed with sulfate. | | Descriptor: | MAGNESIUM ION, Purine nucleoside phosphorylase DeoD-type, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-16 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279 complexed with sulfate.
To be Published
|
|
4M3N
 
 | | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279, NYSGRC Target 029804. | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-06 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279, NYSGRC Target 029804.
To be Published
|
|
4LZA
 
 | | Crystal structure of adenine phosphoribosyltransferase from Thermoanaerobacter pseudethanolicus ATCC 33223, NYSGRC Target 029700. | | Descriptor: | Adenine phosphoribosyltransferase, CHLORIDE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-31 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of adenine phosphoribosyltransferase from Thermoanaerobacter pseudethanolicus ATCC 33223, NYSGRC Target 029700.
To be Published
|
|
7NT7
 
 | |
4V47
 
 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the EF-G.GTP state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
4UV2
 
 | | Structure of the curli transport lipoprotein CsgG in a non-lipidated, pre-pore conformation | | Descriptor: | CURLI PRODUCTION TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
5J99
 
 | | Ambient temperature transition state structure of arginine kinase - crystal 8/Form I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Godsey, M, Davulcu, O, Nix, J, Skalicky, J.J, Bruschweiler, R, Chapman, M.S. | | Deposit date: | 2016-04-08 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Sampling of Conformational Dynamics in Ambient-Temperature Crystal Structures of Arginine Kinase.
Structure, 24, 2016
|
|
4UV3
 
 | | Structure of the curli transport lipoprotein CsgG in its membrane- bound conformation | | Descriptor: | CURLI PRODUCTION ASSEMBLY/TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
6WVR
 
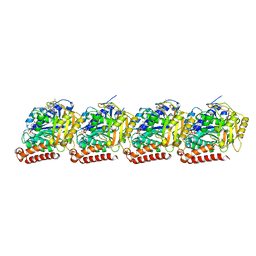 | | Tubulin dimers from a 13-protofilament, Taxol stabilized microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Debs, G.E, Cha, M, Huehn, A.R, Sindelar, C.V. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Dynamic and asymmetric fluctuations in the microtubule wall captured by high-resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WVL
 
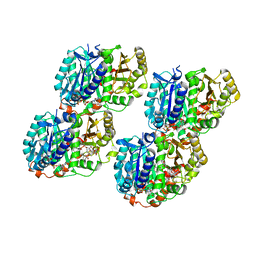 | | Low curvature lateral interaction within a 13-protofilament, Taxol stabilized microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Debs, G.E, Cha, M, Huehn, A.R, Sindelar, C.V. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dynamic and asymmetric fluctuations in the microtubule wall captured by high-resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WVM
 
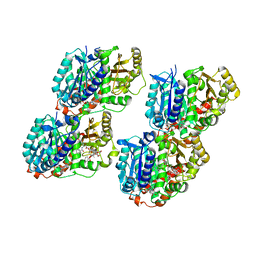 | | High curvature lateral interaction within a 13-protofilament, Taxol stabilized microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Debs, G.E, Cha, M, Huehn, A.R, Sindelar, C.V. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dynamic and asymmetric fluctuations in the microtubule wall captured by high-resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2OWA
 
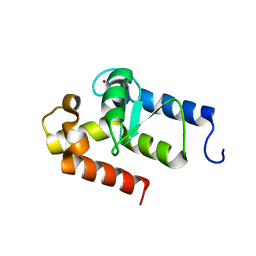 | | Crystal structure of putative GTPase activating protein for ADP ribosylation factor from Cryptosporidium parvum (cgd5_1040) | | Descriptor: | Arfgap-like finger domain containing protein, ZINC ION | | Authors: | Dong, A, Lew, J, Zhao, Y, Hassanali, A, Lin, L, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative GTPase activating protein for ADP ribosylation factor from Cryptosporidium parvum (cgd5_1040)
To be Published
|
|
2P7L
 
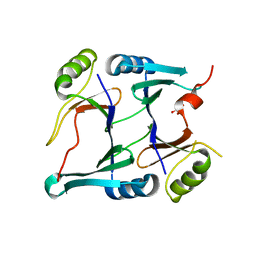 | | Crystal structure of monoclinic form of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes at pH 5.75 | | Descriptor: | GLYCEROL, Glyoxalase family protein | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
2PBF
 
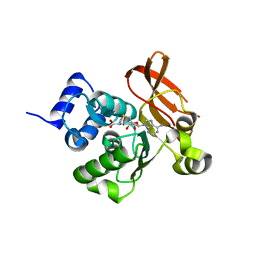 | | Crystal structure of a putative protein-L-isoaspartate O-methyltransferase beta-aspartate methyltransferase (PCMT) from Plasmodium falciparum in complex with S-adenosyl-L-homocysteine | | Descriptor: | Protein-L-isoaspartate O-methyltransferase beta-aspartate methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wernimont, A.K, Hassanali, A, Lin, L, Lew, J, Zhao, Y, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-28 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative protein-L-isoaspartate O-methyltransferase beta-aspartate methyltransferase (PCMT) from Plasmodium falciparum in complex with S-adenosyl-L-homocysteine
To be Published
|
|
2P7P
 
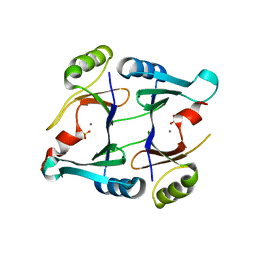 | | Crystal structure of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes complexed with MN(II) and sulfate ion | | Descriptor: | Glyoxalase family protein, MANGANESE (II) ION, SULFATE ION | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
