2HSS
 
 | | 13mer duplex DNA containg an abasic site with beta anomer, averaged structure | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(AAB)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-07-22 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies and molecular modeling of duplex DNA containing normal and 4'-oxidized abasic sites.
Biochemistry, 46, 2007
|
|
2NC3
 
 | | Solution Structure of N-Allosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-D-allopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2KYD
 
 | | RDC and RCSA refinement of an A-form RNA: Improvements in Major Groove Width | | Descriptor: | RNA (5'-R(*CP*UP*AP*GP*UP*UP*AP*GP*CP*UP*AP*AP*CP*UP*AP*G)-3') | | Authors: | Tolbert, B.S, Summers, M.F, Miyazaki, Y, Barton, S, Kinde, B, Stark, P, Singh, R, Bax, A, Case, D. | | Deposit date: | 2010-05-24 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Major groove width variations in RNA structures determined by NMR and impact of 13C residual chemical shift anisotropy and 1H-13C residual dipolar coupling on refinement.
J.Biomol.Nmr, 47, 2010
|
|
1CQO
 
 | |
2NC5
 
 | | Solution Structure of N-Xylosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-D-xylopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2KBU
 
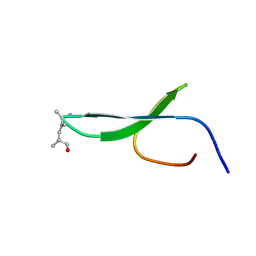 | |
1AX3
 
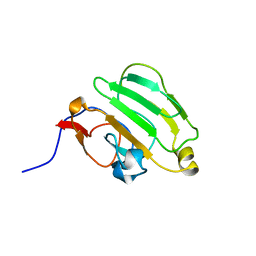 | | SOLUTION NMR STRUCTURE OF B. SUBTILIS IIAGLC, 16 STRUCTURES | | Descriptor: | GLUCOSE PERMEASE IIA DOMAIN | | Authors: | Chen, Y, Case, D.A, Reizer, J, Saier Junior, M.H, Wright, P.E. | | Deposit date: | 1997-10-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of Bacillus subtilis IIAglc.
Proteins, 31, 1998
|
|
1DGQ
 
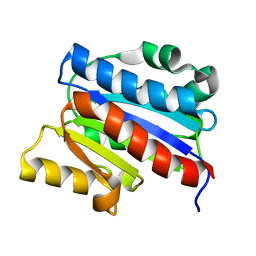 | | NMR SOLUTION STRUCTURE OF THE INSERTED DOMAIN OF HUMAN LEUKOCYTE FUNCTION ASSOCIATED ANTIGEN-1 | | Descriptor: | LEUKOCYTE FUNCTION ASSOCIATED ANTIGEN-1 | | Authors: | Legge, G.B, Kriwacki, R.W, Chung, J, Hommel, U, Ramage, P, Case, D.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 1999-11-24 | | Release date: | 2000-02-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the inserted domain of human leukocyte function associated antigen-1.
J.Mol.Biol., 295, 2000
|
|
2LEF
 
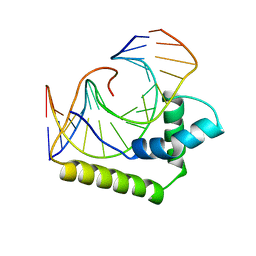 | | LEF1 HMG DOMAIN (FROM MOUSE), COMPLEXED WITH DNA (15BP), NMR, 12 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*CP*CP*CP*TP*TP*TP*GP*AP*AP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*CP*TP*TP*CP*AP*AP*AP*GP*GP*GP*TP*G)-3'), PROTEIN (LYMPHOID ENHANCER-BINDING FACTOR) | | Authors: | Li, X, Love, J.J, Case, D.A, Wright, P.E. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for DNA bending by the architectural transcription factor LEF-1.
Nature, 376, 1995
|
|
2NC6
 
 | | Solution Structure of N-L-idosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-L-idopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
1CNP
 
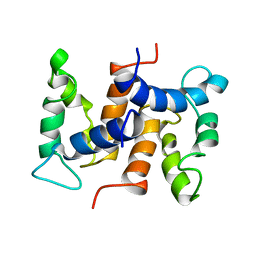 | | THE STRUCTURE OF CALCYCLIN REVEALS A NOVEL HOMODIMERIC FOLD FOR S100 CA2+-BINDING PROTEINS, NMR, 22 STRUCTURES | | Descriptor: | CALCYCLIN (RABBIT, APO) | | Authors: | Potts, B.C.M, Smith, J, Akke, M, Macke, T.J, Okazaki, K, Hidaka, H, Case, D.A, Chazin, W.J. | | Deposit date: | 1995-08-31 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of calcyclin reveals a novel homodimeric fold for S100 Ca(2+)-binding proteins.
Nat.Struct.Biol., 2, 1995
|
|
