6ZMC
 
 | | Structure of the tRNA-Monooxygenase enzyme MiaE frozen under 2000 bar using the high pressure freezing method | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ARGON, CALCIUM ION, ... | | Authors: | Carpentier, P, Atta, M. | | Deposit date: | 2020-07-02 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction.
Nucleic Acids Res., 48, 2020
|
|
6ZMB
 
 | | Structure of the native tRNA-Monooxygenase enzyme MiaE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Carpentier, P, Atta, M. | | Deposit date: | 2020-07-02 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction.
Nucleic Acids Res., 48, 2020
|
|
6ZMA
 
 | | Structure of the tRNA-Monooxygenase enzyme MiaE frozen under 140 bar of krypton using the soak and freeze methodology | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Carpentier, P, Atta, M. | | Deposit date: | 2020-07-02 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction.
Nucleic Acids Res., 48, 2020
|
|
4B30
 
 | |
2WIQ
 
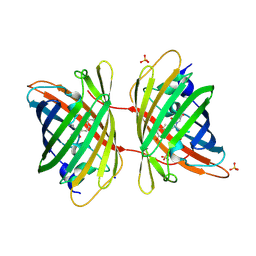 | | Fluorescent protein KillerRed in the native state | | Descriptor: | KILLERRED, SODIUM ION, SULFATE ION | | Authors: | Carpentier, P, Violot, S, Blanchoin, L, Bourgeois, D. | | Deposit date: | 2009-05-15 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Phototoxicity of the Fluorescent Protein Killerred.
FEBS Lett., 583, 2009
|
|
2WIS
 
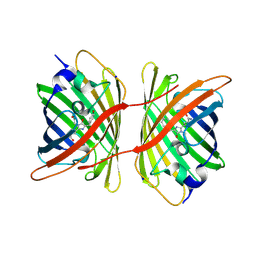 | | Fluorescent protein KillerRed in the bleached state | | Descriptor: | KILLERRED, SODIUM ION | | Authors: | Carpentier, P, Violot, S, Blanchoin, L, Bourgeois, D. | | Deposit date: | 2009-05-15 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Phototoxicity of the Fluorescent Protein Killerred.
FEBS Lett., 583, 2009
|
|
2XBS
 
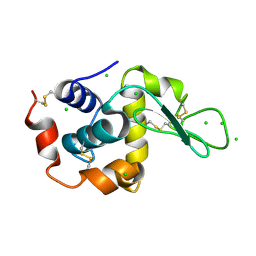 | | Raman crystallography of Hen White Egg Lysozyme - High X-ray dose (16 MGy) | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Carpentier, P, Royant, A, Weik, M, Bourgeois, D. | | Deposit date: | 2010-04-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Raman Assisted Crystallography Reveals a Mechanism of X-Ray Induced Reversible Disulfide Radical Formation
Structure, 18, 2010
|
|
2XBR
 
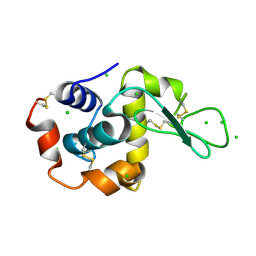 | | Raman crystallography of Hen White Egg Lysozyme - Low X-ray dose (0.2 MGy) | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Carpentier, P, Royant, A, Weik, M, Bourgeois, D. | | Deposit date: | 2010-04-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Raman Assisted Crystallography Reveals a Mechanism of X-Ray Induced Reversible Disulfide Radical Formation
Structure, 18, 2010
|
|
6EYE
 
 | | Crystal structure of murine neuroglobin under 150 bar krypton | | Descriptor: | KRYPTON, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2017-11-12 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping Hydrophobic Tunnels and Cavities in Neuroglobin with Noble Gas under Pressure.
Biophys. J., 113, 2017
|
|
5D51
 
 | | Krypton derivatization of an O2-tolerant membrane-bound [NiFe] hydrogenase reveals a hydrophobic gas tunnel network | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Kalms, J, Schmidt, A, Frielingsdorf, S, van der Linden, P, von Stetten, D, Lenz, O, Carpentier, P, Scheerer, P. | | Deposit date: | 2015-08-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Krypton Derivatization of an O2 -Tolerant Membrane-Bound [NiFe] Hydrogenase Reveals a Hydrophobic Tunnel Network for Gas Transport.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8AFJ
 
 | | tRNA modifying enzyme MiaE soaked in Na-dithionite in a glovebox and flash-cooled using a miniature-airlock | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | van der Linden, P, Engilberge, S, Atta, M, Carpentier, P. | | Deposit date: | 2022-07-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | tRNA modifying enzyme MiaE soaked in Na-dithionite in a glovebox and flash-cooled using a miniature-airlock
To Be Published
|
|
8A9R
 
 | | Tetragonal Hen Egg-White (HEW) Lysozyme soaked in reduced resazurin in a glovebox and flash-cooled using a miniature-airlock | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme | | Authors: | van der Linden, P, Engilberge, S, Atta, M, Carpentier, P. | | Deposit date: | 2022-06-29 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A miniature airlock system to aid the cryo-cooling of protein crystals grown under anoxic conditions
J.Appl.Crystallogr., 2022
|
|
4D7P
 
 | | Superoxide reductase (1Fe-SOR) from Giardia intestinalis | | Descriptor: | FE (III) ION, SUPEROXIDE REDUCTASE | | Authors: | Sousa, C.M, Carpentier, P, Matias, P.M, Testa, F, Pinho, F.G, Sarti, P, Giuffre, A, Bandeiras, T.M, Romao, C.V. | | Deposit date: | 2014-11-26 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Superoxide Reductase from Giardia Intestinalis: Structural Characterization of the First Sor from a Eukaryotic Organism Shows an Iron Centre that is Highly Sensitive to Photoreduction.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3HZ8
 
 | | Crystal structure of the oxidized T176V DsbA1 mutant | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Lafaye, C, Iwema, T, Carpentier, P, Jullian-Binard, C, Serre, L. | | Deposit date: | 2009-06-23 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical and structural study of the homologues of the thiol-disulfide oxidoreductase DsbA in Neisseria meningitidis.
J.Mol.Biol., 392, 2009
|
|
6Q8S
 
 | |
6Q8R
 
 | |
6Q8Q
 
 | |
6QAR
 
 | |
2OXR
 
 | | PAB0955 crystal structure : a GTPase in GDP and Mg bound form from Pyrococcus abyssi (after GTP hydrolysis) | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
1YRA
 
 | | PAB0955 crystal structure : a GTPase in GDP bound form from Pyrococcus abyssi | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
1YR6
 
 | |
1YR8
 
 | | PAB0955 crystal structure : a GTPase in GTP bound form from Pyrococcus abyssi | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
1YR9
 
 | | PAB0955 crystal structure : a GTPase in GDP and PO4 bound form from Pyrococcus abyssi | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
1YR7
 
 | | PAB0955 crystal structure : a GTPase in GTP-gamma-S bound form from Pyrococcus abyssi | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ATP(GTP)binding protein | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
1YRB
 
 | | PAB0955 crystal structure : a GTPase in GDP and Mg bound form from Pyrococcus abyssi | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
