2M58
 
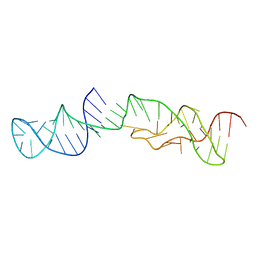 | | Structure of 2'-5' AG1 lariat forming ribozyme in its inactive state | | Descriptor: | RNA (59-MER) | | Authors: | Carlomagno, T, Amata, I, Codutti, L, Falb, M, Fohrer, J, Simon, B. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural principles of RNA catalysis in a 2'-5' lariat-forming ribozyme.
J.Am.Chem.Soc., 135, 2013
|
|
8AOU
 
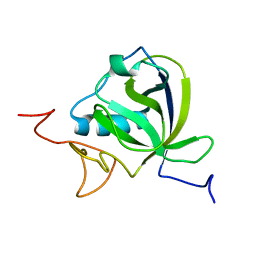 | |
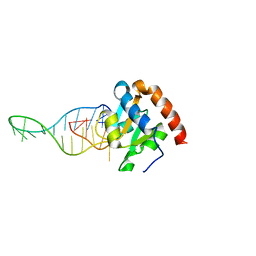
6TPH
 
 | |
6O22
 
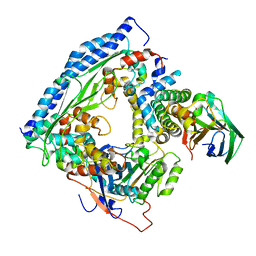 | | Structure of Asf1-H3:H4-Rtt109-Vps75 histone chaperone-lysine acetyltransferase complex with the histone substrate. | | Descriptor: | Histone H3.2, Histone H4, Histone acetyltransferase RTT109, ... | | Authors: | Danilenko, N, Carlomagno, T, Kirkpatrick, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun, 10, 2019
|
|
7OZQ
 
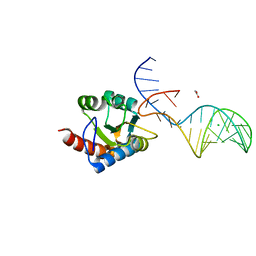 | | Crystal structure of archaeal L7Ae bound to eukaryotic kink-loop | | Descriptor: | 50S ribosomal protein L7Ae, ACETATE ION, CALCIUM ION, ... | | Authors: | Hoefler, S, Lukat, P, Carlomagno, T, Blankenfeldt, W. | | Deposit date: | 2021-06-28 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Eukaryotic Box C/D methylation machinery has two non-symmetric protein assembly sites.
Sci Rep, 11, 2021
|
|
6F0Y
 
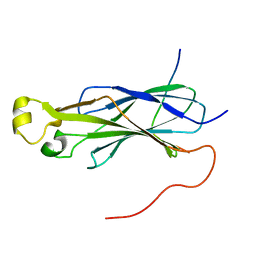 | | Rtt109 peptide bound to Asf1 | | Descriptor: | Histone chaperone ASF1, histone acetyltransferase Rtt109 C-terminus | | Authors: | Lercher, L, Kirkpatrick, J.P, Carlomagno, T. | | Deposit date: | 2017-11-21 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the Asf1-Rtt109 interaction and its role in histone acetylation.
Nucleic Acids Res., 46, 2018
|
|
1OJ5
 
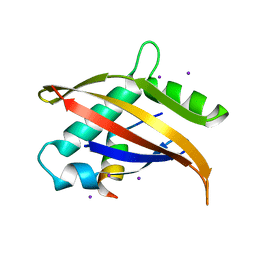 | | Crystal structure of the Nco-A1 PAS-B domain bound to the STAT6 transactivation domain LXXLL motif | | Descriptor: | IODIDE ION, SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 6, STEROID RECEPTOR COACTIVATOR 1A | | Authors: | Razeto, A, Ramakrishnan, V, Giller, K, Lakomek, N, Carlomagno, T, Griesinger, C, Lodrini, M, Litterst, C.M, Pftizner, E, Becker, S. | | Deposit date: | 2003-07-02 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the Ncoa-1/Src-1 Pas-B Domain Bound to the Lxxll Motif of the Stat6 Transactivation Domain
J.Mol.Biol., 336, 2004
|
|
6ZDT
 
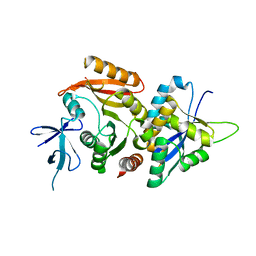 | | Crystal structure of eukaryotic Fibrillarin with Nop56 N-terminal domain | | Descriptor: | Nucleolar protein 56, rRNA 2'-O-methyltransferase fibrillarin | | Authors: | Hoefler, S, Lukat, P, Carlomagno, T, Blankenfeldt, W. | | Deposit date: | 2020-06-15 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | High-resolution structure of eukaryotic Fibrillarin interacting with Nop56 amino-terminal domain.
Rna, 27, 2021
|
|
8QNF
 
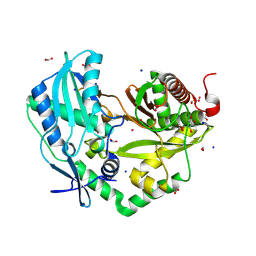 | | Crystal structure of the Condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase | | Descriptor: | Condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase, FORMIC ACID, GLYCEROL, ... | | Authors: | Karanth, M, Schmelz, S, Kirkpatrick, J, Krausze, J, Scrima, A, Carlomagno, T. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The specificity of intermodular recognition in a prototypical nonribosomal peptide synthetase depends on an adaptor domain.
Sci Adv, 10, 2024
|
|
8RZ6
 
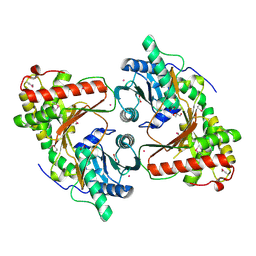 | | SeMet derivative structure of the condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase | | Descriptor: | FORMIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Karanth, M, Schmelz, S, Kirkpatrick, J, Krausze, J, Scrima, A, Carlomagno, T. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The specificity of intermodular recognition in a prototypical nonribosomal peptide synthetase depends on an adaptor domain.
Sci Adv, 10, 2024
|
|
8QPY
 
 | |
8QRX
 
 | |
8QSX
 
 | |
6ROY
 
 | |
7OC3
 
 | |
6ROZ
 
 | |
4BY9
 
 | | The structure of the Box CD enzyme reveals regulation of rRNA methylation | | Descriptor: | 5'-R(*UP*CP*GP*CP*CP*CP*AP*UP*CP*AP*CP)-3', 50S RIBOSOMAL PROTEIN L7AE, FIBRILLARIN-LIKE RRNA/TRNA 2'-O-METHYLTRANSFERASE, ... | | Authors: | Lapinaite, A, Simon, B, Skjaerven, L, Rakwalska-Bange, M, Gabel, F, Carlomagno, T. | | Deposit date: | 2013-07-18 | | Release date: | 2013-10-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Box C/D Enzyme Reveals Regulation of RNA Methylation.
Nature, 502, 2013
|
|
5A17
 
 | |
5A18
 
 | |
4BD3
 
 | |
2XFM
 
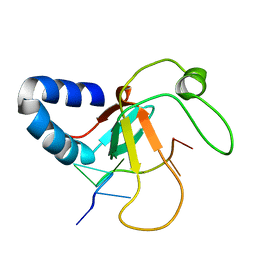 | | Complex structure of the MIWI Paz domain bound to methylated single stranded RNA | | Descriptor: | 5'-R(*AP*CP*CP*GP*AP*CP*UP*(OMU)P)-3', PIWI-LIKE PROTEIN 1 | | Authors: | Simon, B, Kirkpatrick, J.P, Eckhardt, S, Sehr, P, Andrade-Navarro, M.A, Pillai, R.S, Carlomagno, T. | | Deposit date: | 2010-05-27 | | Release date: | 2011-01-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recognition of 2'-O-Methylated 3'-End of Pirna by the Paz Domain of a Piwi Protein.
Structure, 19, 2011
|
|
2XEB
 
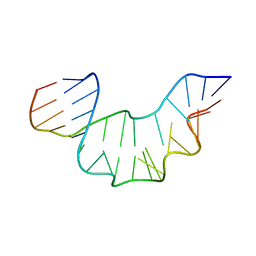 | | NMR STRUCTURE OF THE PROTEIN-UNBOUND SPLICEOSOMAL U4 SNRNA 5' STEM LOOP | | Descriptor: | 5'-R(P*GP*AP*UP*CP*GP*UP*AP*GP*CP*CP*AP*AP*UP*GP*AP* GP*GP*UP*U)-3', 5'-R(P*GP*CP*CP*GP*AP*GP*GP*CP*GP*CP*GP*AP*UP*C)-3' | | Authors: | Falb, M, Amata, I, Gabel, F, Simon, B, Carlomagno, T. | | Deposit date: | 2010-05-12 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the K-turn U4 RNA: a combined NMR and SANS study.
Nucleic Acids Res., 38, 2010
|
|
2KDU
 
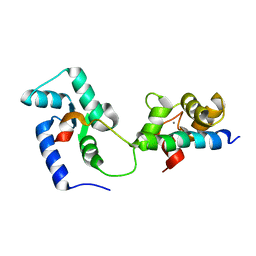 | | Structural basis of the Munc13-1/Ca2+-Calmodulin interaction: A novel 1-26 calmodulin binding motif with a bipartite binding mode | | Descriptor: | CALCIUM ION, Calmodulin, Protein unc-13 homolog A | | Authors: | Rodriguez-Castaneda, F.A, Maestre-Martinez, M, Coudevylle, N, Dimova, K, Jahn, O, Junge, H, Becker, S, Brose, N, Carlomagno, T, Griesinger, C. | | Deposit date: | 2009-01-19 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Modular architecture of Munc13/calmodulin complexes: dual regulation by Ca2+ and possible function in short-term synaptic plasticity.
Embo J., 29, 2010
|
|
2N2P
 
 | | Solution structure of a double base-pair inversion mutant of murine tumour necrosis factor alpha CDE-23 RNA | | Descriptor: | RNA (5'-R(P*GP*CP*AP*UP*GP*UP*UP*UP*AP*GP*UP*GP*UP*CP*UP*AP*AP*AP*CP*GP*GP*UP*U)-3') | | Authors: | Codutti, L, Leppek, K, Zalesak, J, Windeisen, V, Masiewicz, P, Stoecklin, G, Carlomagno, T. | | Deposit date: | 2015-05-11 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Distinct, Sequence-Induced Conformation Is Required for Recognition of the Constitutive Decay Element RNA by Roquin.
Structure, 23, 2015
|
|
2N2O
 
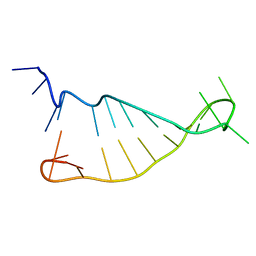 | | Structure of murine tumour necrosis factor alpha CDE RNA | | Descriptor: | RNA (5'-R(P*GP*CP*AP*UP*GP*UP*UP*UP*UP*CP*UP*GP*UP*GP*AP*AP*AP*AP*CP*GP*GP*UP*U)-3') | | Authors: | Codutti, L, Leppek, K, Zalesak, J, Windeisen, V, Masiewicz, P, Stoecklin, G, Carlomagno, T. | | Deposit date: | 2015-05-11 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Distinct, Sequence-Induced Conformation Is Required for Recognition of the Constitutive Decay Element RNA by Roquin.
Structure, 23, 2015
|
|
