2MHC
 
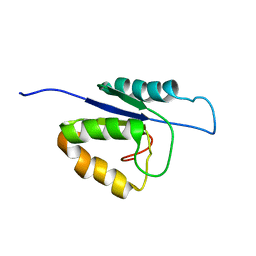 | | NMR structure of the catalytic domain of the large serine resolvase TnpX | | Descriptor: | TnpX | | Authors: | Headey, S.J, Sivakumaran, A, Adams, V, Rodgers, A.J.W, Rood, J.I, Scanlon, M.J, Wilce, M.C.J. | | Deposit date: | 2013-11-20 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and DNA Binding of the Catalytic of the Large Serine Resolvase Tnpx
To be Published
|
|
2M3I
 
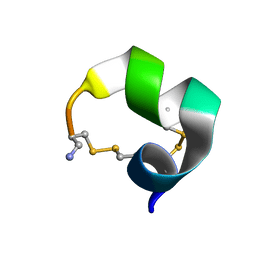 | | Characterization of a Novel Alpha4/6-Conotoxin TxIC from Conus textile that Potently Blocks alpha3beta4 Nicotinic Acetylcholine Receptors | | Descriptor: | Alpha-conotoxin | | Authors: | Luo, S, Zhangsun, D, Zhu, X, Wu, Y, Hu, Y, Christensen, S, Akcan, M, Craik, D.J, McIntosh, J.M. | | Deposit date: | 2013-01-20 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Novel alpha-Conotoxin TxID from Conus textile That Potently Blocks Rat alpha 3 beta 4 Nicotinic Acetylcholine Receptors.
J.Med.Chem., 56, 2013
|
|
1VZM
 
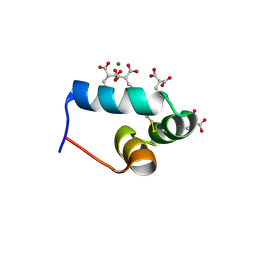 | | OSTEOCALCIN FROM FISH ARGYROSOMUS REGIUS | | Descriptor: | MAGNESIUM ION, OSTEOCALCIN | | Authors: | Frazao, C, Simes, D.C, Coelho, R, Alves, D, Williamson, M.K, Price, P.A, Cancela, M.L, Carrondo, M.A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-09-10 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Evidence of a Fourth Gla Residue in Fish Osteocalcin: Biological Implications
Biochemistry, 44, 2005
|
|
1JC0
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A REDOX-SENSITIVE GREEN FLUORESCENT PROTEIN VARIANT IN A REDUCED FORM | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, Aggeler, R, Oglesbee, D, Cannon, M, Capaldi, R.A, Tsien, R.Y, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators.
J.Biol.Chem., 279, 2004
|
|
1JC1
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A REDOX-SENSITIVE GREEN FLUORESCENT PROTEIN VARIANT IN A OXIDIZED FORM | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, Aggeler, R, Oglesbee, D, Cannon, M, Capaldi, R.A, Tsien, R.Y, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators.
J.Biol.Chem., 279, 2004
|
|
2GD3
 
 | | NMR structure of S14G-humanin in 30% TFE solution | | Descriptor: | Humanin | | Authors: | Benaki, D, Zikos, C, Evangelou, A, Livaniou, E, Vlassi, M, Mikros, E, Pelecanou, M. | | Deposit date: | 2006-03-15 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ser14Gly-humanin, a potent rescue factor against neuronal cell death in Alzheimer's disease.
Biochem.Biophys.Res.Commun., 349, 2006
|
|
2IJY
 
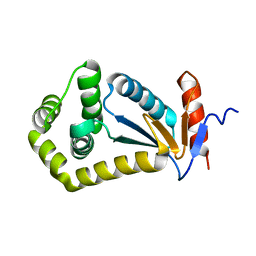 | |
2MQK
 
 | |
2N07
 
 | | Design of a Highly Stable Disulfide-Deleted Mutant of Analgesic Cyclic alpha-Conotoxin Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Yu, R, Seymour, V, Berecki, G, Jia, X, Akcan, M, Adams, D, Kaas, Q, Craik, D. | | Deposit date: | 2015-03-04 | | Release date: | 2016-04-13 | | Method: | SOLUTION NMR | | Cite: | Design of a Highly Stable Disulfide-Deleted Mutant of Analgesic Cyclic alpha-Conotoxin Vc1.1.
To be Published
|
|
1UNF
 
 | | The crystal structure of the eukaryotic FeSOD from Vigna unguiculata suggests a new enzymatic mechanism | | Descriptor: | FE (III) ION, IRON SUPEROXIDE DISMUTASE | | Authors: | Munoz, I.G, Moran, J.F, Becana, M, Montoya, G. | | Deposit date: | 2003-09-10 | | Release date: | 2004-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Crystal Structure of an Eukaryotic Iron Superoxide Dismutase Suggests Intersubunit Cooperation During Catalysis
Protein Sci., 14, 2005
|
|
1LAJ
 
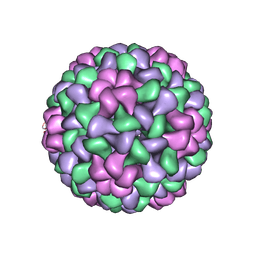 | | The Structure of Tomato Aspermy Virus by X-Ray Crystallography | | Descriptor: | 5'-R(*AP*AP*A)-3', MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Lucas, R.W, Larson, S.B, Canady, M.A, McPherson, A. | | Deposit date: | 2002-03-28 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structure of Tomato Aspermy Virus by X-Ray Crystallography
J.STRUCT.BIOL., 139, 2002
|
|
1MR4
 
 | | Solution Structure of NaD1 from Nicotiana alata | | Descriptor: | Nicotiana alata plant defensin 1 (NaD1) | | Authors: | Lay, F.T, Schirra, H.J, Scanlon, M.J, Anderson, M.A, Craik, D.J. | | Deposit date: | 2002-09-18 | | Release date: | 2003-09-18 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The Three-dimensional Solution Structure of NaD1, a New Floral Defensin from Nicotiana alata and its Application to a Homology Model of the Crop Defense Protein alfAFP
J.MOL.BIOL., 325, 2003
|
|
1Y32
 
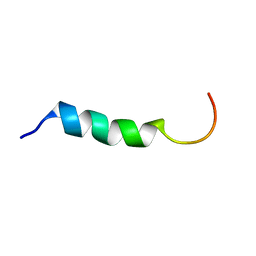 | | NMR structure of humanin in 30% TFE solution | | Descriptor: | Humanin | | Authors: | Benaki, D, Zikos, C, Evangelou, A, Livaniou, E, Vlassi, M, Mikros, E, Pelecanou, M. | | Deposit date: | 2004-11-23 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of humanin, a peptide against Alzheimer's disease-related neurotoxicity.
Biochem.Biophys.Res.Commun., 329, 2005
|
|
2MBS
 
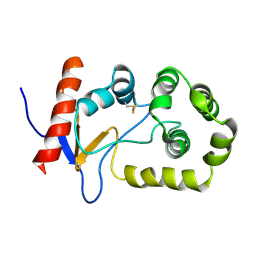 | | NMR solution structure of oxidized KpDsbA | | Descriptor: | Thiol:disulfide interchange protein | | Authors: | Kurth, F, Rimmer, K, Premkumar, L, Mohanty, B, Duprez, W, Halili, M.A, Shouldice, S.R, Heras, B, Fairlie, D.P, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2013-08-03 | | Release date: | 2013-12-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comparative Sequence, Structure and Redox Analyses of Klebsiella pneumoniae DsbA Show That Anti-Virulence Target DsbA Enzymes Fall into Distinct Classes.
Plos One, 8, 2013
|
|
2JWD
 
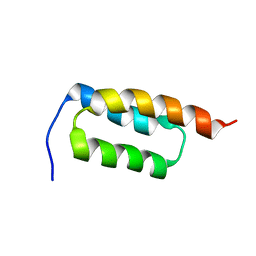 | | protein A | | Descriptor: | Immunoglobulin G-binding protein A | | Authors: | Robertson, A, Horne, J, Scanlon, M.J, Bottomley, S.P. | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Polyglutamine length-dependent midfolding is confined to the Poly-Q region
To be Published
|
|
1LDL
 
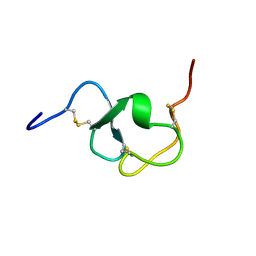 | |
1QH2
 
 | |
2M32
 
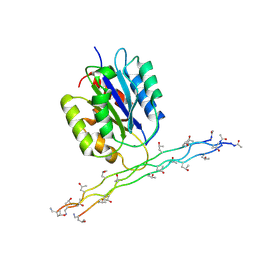 | | Alpha-1 integrin I-domain in complex with GLOGEN triple helical peptide | | Descriptor: | GLOGEN peptide, Integrin alpha-1, MAGNESIUM ION | | Authors: | Chin, Y, Headey, S, Mohanty, B, McEwan, P, Swarbrick, J, Mulhern, T, Emsley, J, Simpson, J, Scanlon, M. | | Deposit date: | 2013-01-07 | | Release date: | 2013-11-06 | | Last modified: | 2014-02-12 | | Method: | SOLUTION NMR | | Cite: | The Structure of Integrin alpha 1I Domain in Complex with a Collagen-mimetic Peptide.
J.Biol.Chem., 288, 2013
|
|
2KB4
 
 | | NMR structure of the unphosphorylated form of OdhI, OdhI. | | Descriptor: | Oxoglutarate dehydrogenase inhibitor | | Authors: | Barthe, P, Roumestand, C, Canova, M, Hurard, C, Molle, V, Cohen-Gonsaud, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Dynamic and Structural Characterization of a Bacterial FHA Protein Reveals a New Autoinhibition Mechanism.
Structure, 17, 2009
|
|
2KB3
 
 | | NMR Structure of the phosphorylated form of OdhI, pOdhI. | | Descriptor: | Oxoglutarate dehydrogenase inhibitor | | Authors: | Barthe, P, Roumestand, C, Canova, M, Hurard, C, Molle, V, Cohen-Gonsaud, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Dynamic and Structural Characterization of a Bacterial FHA Protein Reveals a New Autoinhibition Mechanism.
Structure, 17, 2009
|
|
4BYY
 
 | | Apo GlxR | | Descriptor: | GLYCEROL, PHOSPHATE ION, TRANSCRIPTIONAL REGULATOR, ... | | Authors: | Pohl, E, Cann, M.J, Townsend, P.D, Money, V.A. | | Deposit date: | 2013-07-21 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The Crystal Structures of Apo and Camp-Bound Glxr from Corynebacterium Glutamicum Reveal Structural and Dynamic Changes Upon Camp Binding in Crp/Fnr Family Transcription Factors.
Plos One, 9, 2014
|
|
4BS1
 
 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR (NTRC FAMILY) | | Authors: | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AHT
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
4AHS
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1-BENZOFURAN-7-CARBOXYLIC ACID, ACETATE ION, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
4AH9
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-PHENYL-1,2,4-THIADIAZOL-5-YL)-1,4-DIAZEPANE, CHLORIDE ION, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-06 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
