3VQC
 
 | | HIV-1 IN core domain in complex with (5-METHYL-3-PHENYL-1,2-OXAZOL-4-YL)METHANOL | | Descriptor: | (5-methyl-3-phenyl-1,2-oxazol-4-yl)methanol, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQD
 
 | | HIV-1 IN core domain in complex with 5-methyl-3-phenyl-1,2-oxazole-4-carboxylic acid | | Descriptor: | 5-methyl-3-phenyl-1,2-oxazole-4-carboxylic acid, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQ9
 
 | | HIV-1 IN core domain in complex with 6-fluoro-1,3-benzothiazol-2-amine | | Descriptor: | 6-fluoro-1,3-benzothiazol-2-amine, POL polyprotein | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQP
 
 | | HIV-1 IN core domain in complex with 2,3-dihydro-1,4-benzodioxin-5-ylmethanol | | Descriptor: | 2,3-dihydro-1,4-benzodioxin-5-ylmethanol, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-29 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
4JCG
 
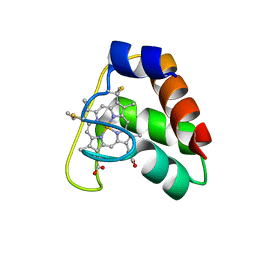 | | Recombinant wild type Nitrosomonas europaea cytochrome c552 | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Wedekind, J.E, Can, M, Krucinska, J, Bren, K.L. | | Deposit date: | 2013-02-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Characterization of Nitrosomonas europaea Cytochrome c-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
5C4I
 
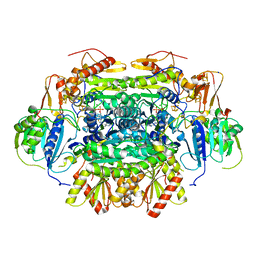 | | Structure of an Oxalate Oxidoreductase | | Descriptor: | IRON/SULFUR CLUSTER, MAGNESIUM ION, Oxalate oxidoreductase subunit alpha, ... | | Authors: | Gibson, M.I, Brignole, E.J, Pierce, E, Can, M, Ragsdale, S.W, Drennan, C.L. | | Deposit date: | 2015-06-18 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.274 Å) | | Cite: | The Structure of an Oxalate Oxidoreductase Provides Insight into Microbial 2-Oxoacid Metabolism.
Biochemistry, 54, 2015
|
|
6VLD
 
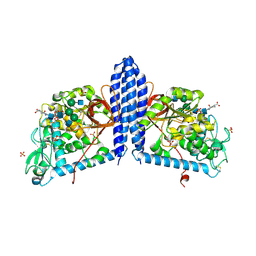 | | Crystal structure of human alpha 1,6-fucosyltransferase, FUT8 bound to GDP and A2SGP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASPARAGINE, Alpha-(1,6)-fucosyltransferase, ... | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E.D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
6VGF
 
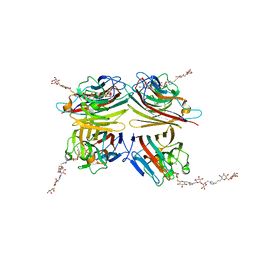 | | Peanut lectin complexed with divalent S-beta-D-thiogalactopyranosyl beta-D-glucopyranoside derivative (diSTGD) | | Descriptor: | (2S,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5S,6S)-3,4,5-trihydroxy-6-({[(1-{[(2R,3S,4S,5R,6R)-3,4,5-trihydroxy-6-{[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-({4-[({[(2S,3S,4S,5R,6S)-3,4,5-trihydroxy-6-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-2-yl]methyl}sulfanyl)methyl]-1H-1,2,3-triazol-1-yl}methyl)tetrahydro-2H-pyran-2-yl]oxy}tetrahydro-2H-pyran-2-yl]methyl}-1H-1,2,3-triazol-4-yl)methyl]sulfanyl}methyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-3,4,5-triol, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VC4
 
 | | Peanut lectin complexed with S-beta-D-Thiogalactopyranosyl beta-D-glucopyranoside derivative (STGD) | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5S,6S)-3,4,5-trihydroxy-6-({[(1-{[(2R,3S,4S,5R,6S)-3,4,5-trihydroxy-6-methoxytetrahydro-2H-pyran-2-yl]methyl}-1H-1,2,3-triazol-4-yl)methyl]sulfanyl}methyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-3,4,5-triol (non-preferred name), CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VC3
 
 | | Peanut lectin complexed with S-beta-D-thiogalactopyranosyl 6-deoxy-6-S-propynyl-beta-D-glucopyranoside (STG) | | Descriptor: | 6-S-(prop-2-yn-1-yl)-6-thio-beta-D-glucopyranosyl 1-thio-beta-D-galactopyranoside, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VLF
 
 | | Crystal structure of mouse alpha 1,6-fucosyltransferase, FUT8 in its Apo-form | | Descriptor: | 1,2-ETHANEDIOL, Alpha-(1,6)-fucosyltransferase, SULFATE ION | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
6VLG
 
 | | Crystal structure of mouse alpha 1,6-fucosyltransferase, FUT8 bound to GDP | | Descriptor: | Alpha-(1,6)-fucosyltransferase, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E.D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
6VLE
 
 | | Crystal structure of human alpha 1,6-fucosyltransferase, FUT8 in its Apo-form | | Descriptor: | Alpha-(1,6)-fucosyltransferase, SULFATE ION | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E.D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
6VUC
 
 | |
6VUJ
 
 | | Crystal structure of BRD4 bromodomain 1 with N-methylpyrrolidin-2-one (NMP) derivative 15c (N,N-diethyl-3',4'-dimethoxy-6-(1-methyl-5-oxopyrrolidin-3-yl)-[1,1'-biphenyl]-3-sulfonamide) | | Descriptor: | Bromodomain-containing protein 4, N,N-diethyl-3',4'-dimethoxy-6-[(3S)-1-methyl-5-oxopyrrolidin-3-yl][1,1'-biphenyl]-3-sulfonamide, NITRATE ION | | Authors: | Ilyichova, O.V, Scanlon, M.J, Thompson, P.E. | | Deposit date: | 2020-02-15 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Substituted 1-methyl-4-phenylpyrrolidin-2-ones - Fragment-based design of N-methylpyrrolidone-derived bromodomain inhibitors.
Eur.J.Med.Chem., 191, 2020
|
|
6VUB
 
 | |
6VUF
 
 | |
3ZOW
 
 | | Crystal Structure of Wild Type Nitrosomonas europaea Cytochrome c552 | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Karlsen, S, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-25 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
7S1C
 
 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001897 (compound 1) | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, ~{N}-methyl-1-(3-thiophen-3-ylphenyl)methanamine | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Sharma, P. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
7S1L
 
 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001896 (compound 72) | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, methyl cis-4-({[3-(thiophen-3-yl)benzyl]amino}methyl)cyclohexanecarboxylate | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Caria, S. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
7S1D
 
 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001877 (compound 39) | | Descriptor: | 1-[3-(thiophen-3-yl)benzyl]piperidin-2-one, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Caria, S. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
7S1F
 
 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001886 (compound 38) | | Descriptor: | 1-[(3-thiophen-3-ylphenyl)methyl]-3~{H}-pyrrol-2-one, COPPER (II) ION, GLYCEROL, ... | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Caria, S. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
3ZOY
 
 | | Crystal Structure of N64Del Mutant of Nitrosomonas europaea Cytochrome c552 (hexagonal space group) | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
3ZOX
 
 | | Crystal Structure of N64Del Mutant of Nitrosomonas europaea Cytochrome c552 (monoclinic space group) | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
6PLI
 
 | | Crystal Structure of EcDsbA in a complex with purified oxadiazole 11 | | Descriptor: | 2-[4-(4-cyano-3-methylphenoxy)phenyl]-N-methyl-N-[2-(5-methyl-1,2,4-oxadiazol-3-yl)ethyl]acetamide, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-07-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
