6GQP
 
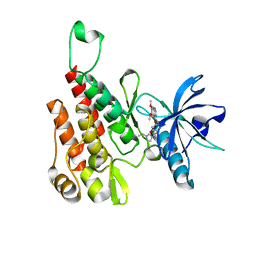 | | Crystal structure of human KDR (VEGFR2) kinase domain in complex with AZD3229-analogue (compound 23) | | Descriptor: | Vascular endothelial growth factor receptor 2, ~{N}-[4-(6,7-dimethoxyquinazolin-4-yl)oxyphenyl]-2-(1-ethylpyrazol-4-yl)ethanamide | | Authors: | Hardy, C.J, Schimpl, M, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
6ODI
 
 | | Structure of CagY from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagY | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6GQM
 
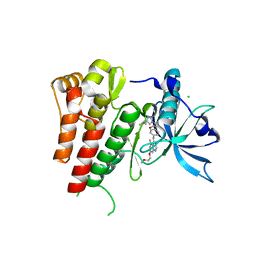 | | Crystal structure of human c-KIT kinase domain in complex with a small molecule inhibitor, AZD3229 | | Descriptor: | CHLORIDE ION, Mast/stem cell growth factor receptor Kit, ~{N}-[4-[[5-fluoranyl-7-(2-methoxyethoxy)quinazolin-4-yl]amino]phenyl]-2-(4-propan-2-yl-1,2,3-triazol-1-yl)ethanamide | | Authors: | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
8U1W
 
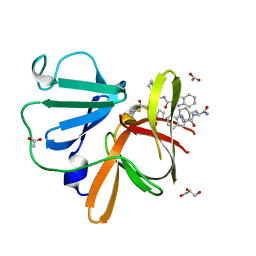 | | Structure of Norovirus (Hu/GII.4/Sydney/NSW0514/2012/AU) protease bound to inhibitor NV-004 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidase C37, ... | | Authors: | Eruera, A.R, Campbell, A.C, Krause, K.L. | | Deposit date: | 2023-09-03 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of Inhibitor-Bound GII.4 Sydney 2012 Norovirus 3C-Like Protease.
Viruses, 15, 2023
|
|
8U1V
 
 | |
6WPU
 
 | |
6X0H
 
 | |
6X0J
 
 | |
6X0I
 
 | |
6X9A
 
 | |
7JVK
 
 | |
6X9B
 
 | |
6X9C
 
 | |
6X99
 
 | |
6X9D
 
 | |
6OEH
 
 | | PolyAla Model of OMCC I-Layer | | Descriptor: | PolyAla Model of OMCC I-Layer | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
7JVL
 
 | |
6OEF
 
 | | PolyAla Model of the O-layer from the Type 4 Secretion System of H. pylori | | Descriptor: | PolyAla Model of OMCC O-Layer | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
2C2M
 
 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-14-[4-(BENZYLOXY)-4-OXOBUTANOYL]-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14 -PENTAAZAHEXADECAN-16-OIC ACID, ... | | Authors: | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A, Mikolajczyk, J, Salvesen, G.S, Gruetter, M.G, Powers, J.C. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
2C2O
 
 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-14-{4-[BENZYL(METHYL) AMINO]-4-OXOBUTANOYL}-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN-16-OIC ACID, ... | | Authors: | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A, Mikolajczyk, J, Salvesen, G.S, Gruetter, M.G, Powers, J.C. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
6OEG
 
 | | Structure of CagX from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagX | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6ODJ
 
 | | PolyAla Model of the PRC from the Type 4 Secretion System of H. pylori | | Descriptor: | PolyAla Model of PRC from H.pylori | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
1XOB
 
 | | THIOREDOXIN (REDUCED DITHIO FORM), NMR, 20 STRUCTURES | | Descriptor: | THIOREDOXIN | | Authors: | Jeng, M.-F, Campbell, A.P, Begley, T, Holmgren, A, Case, D.A, Wright, P.E, Dyson, H.J. | | Deposit date: | 1995-11-28 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin.
Structure, 2, 1994
|
|
1XOA
 
 | | THIOREDOXIN (OXIDIZED DISULFIDE FORM), NMR, 20 STRUCTURES | | Descriptor: | THIOREDOXIN | | Authors: | Jeng, M.-F, Campbell, A.P, Begley, T, Holmgren, A, Case, D.A, Wright, P.E, Dyson, H.J. | | Deposit date: | 1995-11-28 | | Release date: | 1996-06-10 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin.
Structure, 2, 1994
|
|
1TW9
 
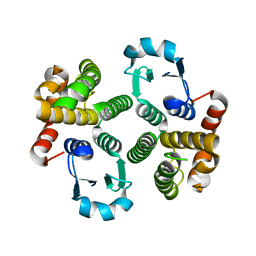 | | Glutathione Transferase-2, apo form, from the nematode Heligmosomoides polygyrus | | Descriptor: | glutathione S-transferase 2 | | Authors: | Schuller, D.J, Liu, Q, Kriksunov, I.A, Campbell, A.M, Barrett, J, Brophy, P.M, Hao, Q. | | Deposit date: | 2004-06-30 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of a new class of glutathione transferase from the model human hookworm nematode Heligmosomoides polygyrus.
Proteins, 61, 2005
|
|
