1MIT
 
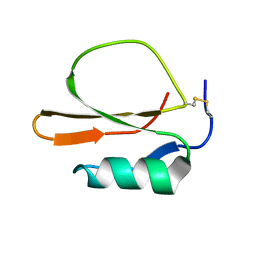 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | 分子名称: | TRYPSIN INHIBITOR V | | 著者 | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | 登録日 | 1995-10-26 | | 公開日 | 1996-04-03 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
1CCH
 
 | |
1COR
 
 | |
1HYM
 
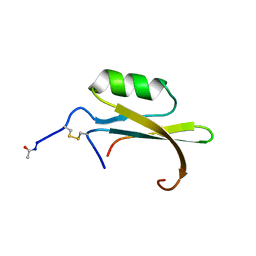 | | HYDROLYZED TRYPSIN INHIBITOR (CMTI-V, MINIMIZED AVERAGE NMR STRUCTURE) | | 分子名称: | HYDROLYZED CUCURBITA MAXIMA TRYPSIN INHIBITOR V | | 著者 | Cai, M, Gong, Y, Prakash, O, Krishnamoorthi, R. | | 登録日 | 1995-06-12 | | 公開日 | 1995-09-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Reactive-site hydrolyzed Cucurbita maxima trypsin inhibitor-V: function, thermodynamic stability, and NMR solution structure.
Biochemistry, 34, 1995
|
|
1WJF
 
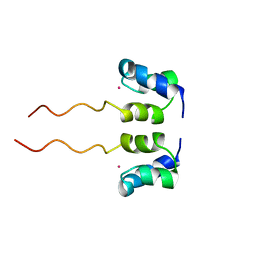 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, 40 STRUCTURES | | 分子名称: | CADMIUM ION, HIV-1 INTEGRASE | | 著者 | Cai, M, Gronenborn, A.M, Clore, G.M. | | 登録日 | 1998-06-11 | | 公開日 | 1998-12-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
6U6P
 
 | |
6U6R
 
 | |
6U6S
 
 | |
6U6Q
 
 | |
1TIN
 
 | |
1WJE
 
 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | CADMIUM ION, HIV-1 INTEGRASE | | 著者 | Cai, M, Gronenborn, A.M, Clore, G.M. | | 登録日 | 1998-06-11 | | 公開日 | 1998-12-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
2LRK
 
 | | Solution Structures of the IIA(Chitobiose)-HPr complex of the N,N'-Diacetylchitobiose | | 分子名称: | N,N'-diacetylchitobiose-specific phosphotransferase enzyme IIA component, Phosphocarrier protein HPr | | 著者 | Cai, M, Jung, Y, Clore, M. | | 登録日 | 2012-04-06 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the IIAChitobiose-HPr Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia coli Phosphotransferase System.
J.Biol.Chem., 287, 2012
|
|
2LOE
 
 | | Structure of the Plasmodium 6-cysteine s48/45 Domain | | 分子名称: | 6-cysteine protein, putative | | 著者 | Cai, M, Arredondo, S.A, Clore, M.G, Miller, L.H, Takayama, Y, Macdonald, N.J, Enderson, E.D, Aravind, L. | | 登録日 | 2012-01-23 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the Plasmodium 6-cysteine s48/45 domain.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1F9X
 
 | | AVERAGE NMR SOLUTION STRUCTURE OF THE BIR-3 DOMAIN OF XIAP | | 分子名称: | INHIBITOR OF APOPTOSIS PROTEIN XIAP, ZINC ION | | 著者 | Sun, C, Cai, M, Meadows, R.P, Fesik, S.W. | | 登録日 | 2000-07-11 | | 公開日 | 2001-07-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure and mutagenesis of the third Bir domain of the inhibitor of apoptosis protein XIAP.
J.Biol.Chem., 275, 2000
|
|
1GJJ
 
 | |
1WJA
 
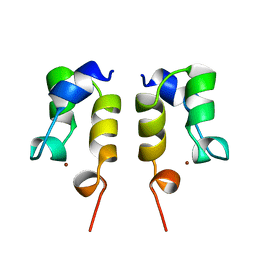 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, REGULARIZED MEAN STRUCTURE | | 分子名称: | HIV-1 INTEGRASE, ZINC ION | | 著者 | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | 登録日 | 1997-05-13 | | 公開日 | 1998-05-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJD
 
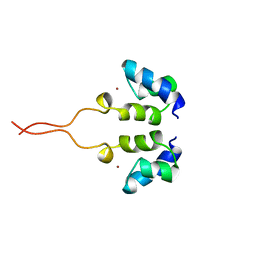 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (E FORM), NMR, 38 STRUCTURES | | 分子名称: | HIV-1 INTEGRASE, ZINC ION | | 著者 | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | 登録日 | 1997-05-13 | | 公開日 | 1998-05-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJB
 
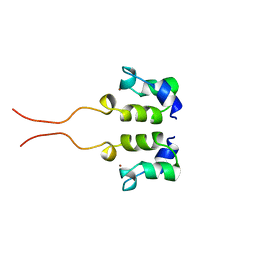 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, 40 STRUCTURES | | 分子名称: | HIV-1 INTEGRASE, ZINC ION | | 著者 | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | 登録日 | 1997-05-13 | | 公開日 | 1998-05-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJC
 
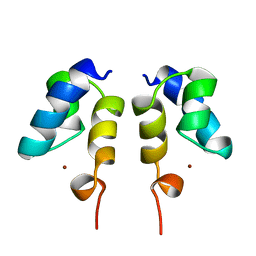 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (E FORM), NMR, REGULARIZED MEAN STRUCTURE | | 分子名称: | HIV-1 INTEGRASE, ZINC ION | | 著者 | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | 登録日 | 1997-05-13 | | 公開日 | 1998-05-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
6FCE
 
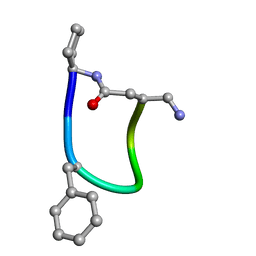 | | NMR ensemble of Macrocyclic Peptidomimetic Containing Constrained a,a-dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors | | 分子名称: | ACP-HIS-DPHE-ARG-TRP-ASP-NH2 | | 著者 | Brancaccio, D, Carotenuto, A, Grieco, P, Merlino, F, Zhou, Y, Cai, M, Yousif, A.M, Di Maro, S, Novellino, E, Hruby, V.J. | | 登録日 | 2017-12-20 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Development of Macrocyclic Peptidomimetics Containing Constrained alpha , alpha-Dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors.
J. Med. Chem., 61, 2018
|
|
1ZHQ
 
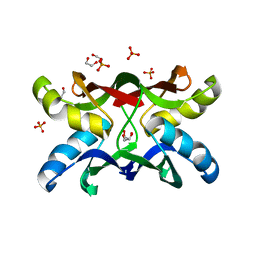 | | Crystal structure of apo MVL | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, mannan-binding lectin | | 著者 | Williams, D.C, Lee, J.Y, Cai, M, Bewley, C.A, Clore, G.M. | | 登録日 | 2005-04-26 | | 公開日 | 2005-06-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structures of the HIV-1 Inhibitory Cyanobacterial Protein MVL Free and Bound to Man3GlcNAc2: STRUCTURAL BASIS FOR SPECIFICITY AND HIGH-AFFINITY BINDING TO THE CORE PENTASACCHARIDE FROM N-LINKED OLIGOMANNOSIDE.
J.Biol.Chem., 280, 2005
|
|
1ZHS
 
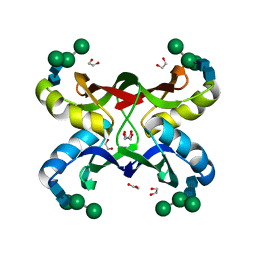 | | Crystal structure of MVL bound to Man3GlcNAc2 | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Williams, D.C, Lee, J.Y, Cai, M, Bewley, C.A, Clore, G.M. | | 登録日 | 2005-04-26 | | 公開日 | 2005-06-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of the HIV-1 Inhibitory Cyanobacterial Protein MVL Free and Bound to Man3GlcNAc2: STRUCTURAL BASIS FOR SPECIFICITY AND HIGH-AFFINITY BINDING TO THE CORE PENTASACCHARIDE FROM N-LINKED OLIGOMANNOSIDE.
J.Biol.Chem., 280, 2005
|
|
1VKR
 
 | |
2ODG
 
 | |
1VRV
 
 | |
