5CPL
 
 | | The crystal structure of Xenobiotic reductase A (XenA) from Pseudomonas putida in complex with a nicotinamide mimic (mNH2) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 1-benzyl-1,4,5,6-tetrahydropyridine-3-carboxamide, CALCIUM ION, ... | | Authors: | Knaus, T, Paul, C.E, Levy, C.W, Mutti, F.G, Hollmann, F, Scrutton, N.S. | | Deposit date: | 2015-07-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Better than Nature: Nicotinamide Biomimetics That Outperform Natural Coenzymes.
J.Am.Chem.Soc., 138, 2016
|
|
5CPN
 
 | | Crystal structure of XenA from Pseudomonas putida in complex with an NADH mimic (mAc) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 1-[(3S)-1-benzylpiperidin-3-yl]ethanone, Xenobiotic reductase | | Authors: | Knaus, T, Paul, C.E, Levy, C.W, Mutti, F.G, Hollmann, F, Scrutton, N.S. | | Deposit date: | 2015-07-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Better than Nature: Nicotinamide Biomimetics That Outperform Natural Coenzymes.
J.Am.Chem.Soc., 138, 2016
|
|
5BRV
 
 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | Carbonic anhydrase 2, ZINC ION, pentamethylcyclopentadienyl iridium [N-benzensulfonamide-(2-pyridylmethyl-4-benzensulfonamide)amin] chloride | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
5BRU
 
 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | Carbonic anhydrase 2, SULFATE ION, ZINC ION, ... | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
5CPM
 
 | | XenA from Pseudomonas putida in complex with NADPH4. | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Xenobiotic reductase | | Authors: | Knaus, T, Paul, C.E, Levy, C.W, Mutti, F.G, Hollmann, F, Scrutton, N.S. | | Deposit date: | 2015-07-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Better than Nature: Nicotinamide Biomimetics That Outperform Natural Coenzymes.
J.Am.Chem.Soc., 138, 2016
|
|
5BRW
 
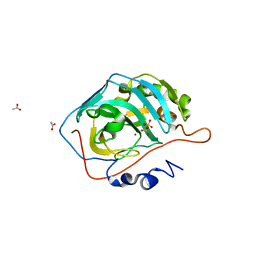 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | ACETATE ION, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
1MZF
 
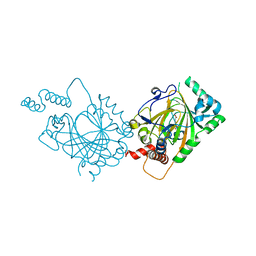 | |
1NDU
 
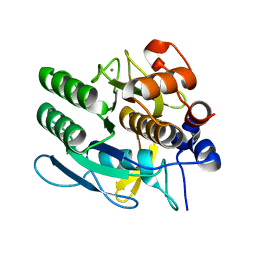 | |
7KPV
 
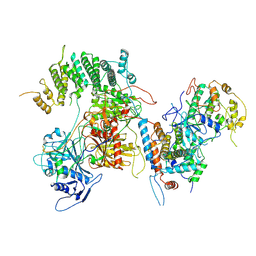 | | Structure of kinase and Central lobes of yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12, Mediator of RNA polymerase II transcription subunit 13, Meiotic mRNA stability protein kinase SSN3, ... | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
1PGP
 
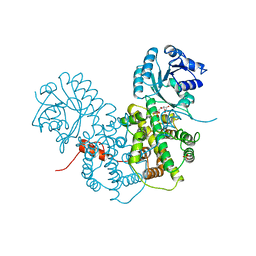 | | CRYSTALLOGRAPHIC STUDY OF COENZYME, COENZYME ANALOGUE AND SUBSTRATE BINDING IN 6-PHOSPHOGLUCONATE DEHYDROGENASE: IMPLICATIONS FOR NADP SPECIFICITY AND THE ENZYME MECHANISM | | Descriptor: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, 6-PHOSPHOGLUCONIC ACID, SULFATE ION | | Authors: | Adams, M.J, Phillips, C, Gover, S, Naylor, C.E. | | Deposit date: | 1994-07-18 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism.
Structure, 2, 1994
|
|
7KGE
 
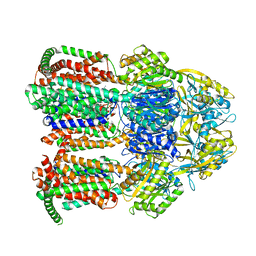 | |
7KGI
 
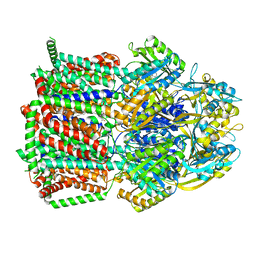 | |
7KGF
 
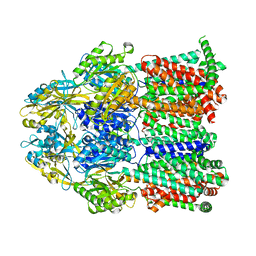 | |
7KGH
 
 | |
7KGD
 
 | |
7KGG
 
 | |
1NN4
 
 | | Structural Genomics, RpiB/AlsB | | Descriptor: | Ribose 5-phosphate isomerase B | | Authors: | Zhang, R.G, Andersson, C.E, Mowbray, S.L, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S.L, Arrowsmith, C, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-12 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ribose-5-phosphate isomerase reaction.
J.Mol.Biol., 332, 2003
|
|
1Q07
 
 | | Crystal structure of the Au(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | GOLD ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
7JT1
 
 | |
7JZ2
 
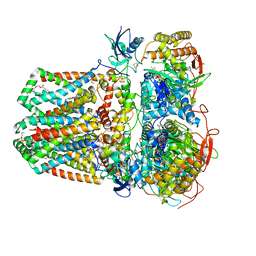 | | Succinate: quinone oxidoreductase SQR from E.coli K12 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lyu, M, Su, C.-C, Morgan, C.E, Bolla, J.R, Robinson, C.V, Yu, E.W. | | Deposit date: | 2020-09-01 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
7JSS
 
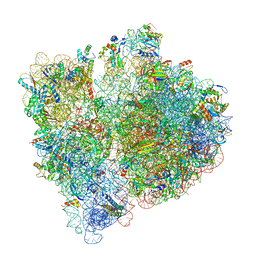 | |
1MWY
 
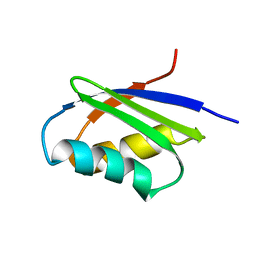 | | Solution structure of the N-terminal domain of ZntA in the apo-form | | Descriptor: | ZntA | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Finney, L.A, Outten, C.E, O'Halloran, T.V. | | Deposit date: | 2002-10-01 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new zinc-protein coordination site in intracellular metal trafficking: solution structure of the apo and Zn(II) forms of ZntA (46-118)
J.Mol.Biol., 323, 2002
|
|
7JSZ
 
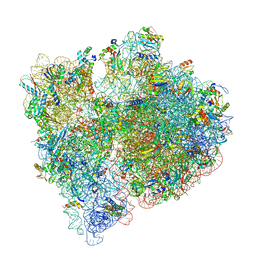 | |
1MWZ
 
 | | Solution structure of the N-terminal domain of ZntA in the Zn(II)-form | | Descriptor: | ZINC ION, ZntA | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Finney, L.A, Outten, C.E, O'Halloran, T.V. | | Deposit date: | 2002-10-01 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new zinc-protein coordination site in intracellular metal trafficking: solution structure of the apo and Zn(II) forms of ZntA (46-118)
J.Mol.Biol., 323, 2002
|
|
7K7K
 
 | |
