6BUW
 
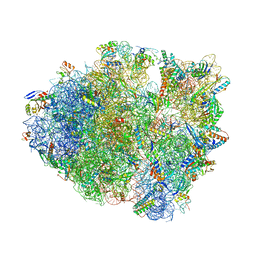 | | Thermus thermophilus 70S complex containing 16S G299A ram mutation and empty A site. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2017-12-11 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding.
Nucleic Acids Res., 47, 2019
|
|
7QYR
 
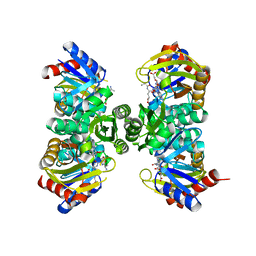 | | Crystal structure of RimK from Pseudomonas aeruginosa PAO1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Probable alpha-L-glutamate ligase, poly-glutamate | | Authors: | Thompson, C.M.A, Little, R.H, Stevenson, C.E.M, Lawson, D.M, Malone, J.G. | | Deposit date: | 2022-01-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the mechanism of adaptive ribosomal modification by Pseudomonas RimK.
Proteins, 91, 2023
|
|
7QYS
 
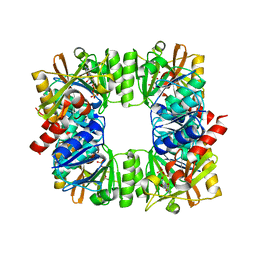 | | Crystal structure of RimK from Pseudomonas syringae DC3000 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Probable alpha-L-glutamate ligase | | Authors: | Thompson, C.M.A, Little, R.H, Stevenson, C.E.M, Lawson, D.M, Malone, J.G. | | Deposit date: | 2022-01-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of adaptive ribosomal modification by Pseudomonas RimK.
Proteins, 91, 2023
|
|
5UFW
 
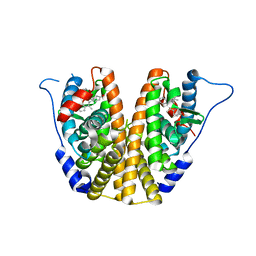 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with OP1154 | | Descriptor: | (2S)-3-(4-hydroxyphenyl)-4-methyl-2-(4-{2-[(3S)-3-methylpyrrolidin-1-yl]ethoxy}phenyl)-2H-1-benzopyran-7-ol, Estrogen receptor | | Authors: | Fanning, S.W, Hodges-Gallagher, L, Myles, D.C, Sun, R, Fowler, C.E, Green, B.D, Harmon, C.L, Greene, G.L, Kushner, P.J. | | Deposit date: | 2017-01-06 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Specific stereochemistry of OP-1074 disrupts estrogen receptor alpha helix 12 and confers pure antiestrogenic activity.
Nat Commun, 9, 2018
|
|
7QOA
 
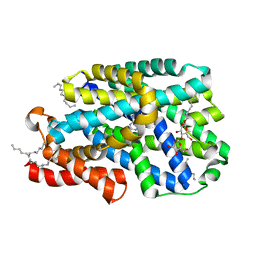 | | Structure of CodB, a cytosine transporter in an outward-facing conformation | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-AMINOPYRIMIDIN-2(1H)-ONE, Cytosine permease, ... | | Authors: | Hatton, C.E, Cameron, A.D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of cytosine transport protein CodB provides insight into nucleobase-cation symporter 1 mechanism.
Embo J., 41, 2022
|
|
3TKZ
 
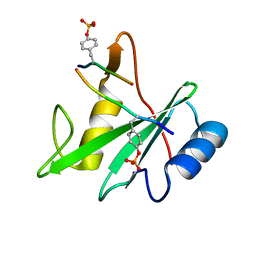 | | Structure of the SHP-2 N-SH2 domain in a 1:2 complex with RVIpYFVPLNR peptide | | Descriptor: | PROTEIN (RVIpYFVPLNR peptide), Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Y, Zhang, J, Yuan, C, Hard, R.L, Park, I.H, Li, C, Bell, C.E, Pei, D. | | Deposit date: | 2011-08-29 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Simultaneous binding of two peptidyl ligands by a SRC homology 2 domain.
Biochemistry, 50, 2011
|
|
4CN1
 
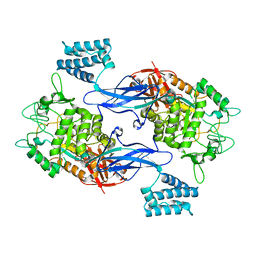 | | GlgE isoform 1 from Streptomyces coelicolor D394A mutant with maltose- 1-phosphate bound | | Descriptor: | ALPHA-1,4-GLUCAN: MALTOSE-1-PHOSPHATE MALTOSYLTRANSFERASE 1, alpha-D-glucopyranose-(1-4)-1-O-phosphono-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rashid, A.M, Saalbach, G, Tang, M, Tuukanen, A, Svergun, D.I, Withers, S.G, Lawson, D.M, Bornemann, S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insight Into How Streptomyces Coelicolor Maltosyl Transferase Glge Binds Alpha-Maltose 1-Phosphate and Forms a Maltosyl-Enzyme Intermediate.
Biochemistry, 53, 2014
|
|
3TL0
 
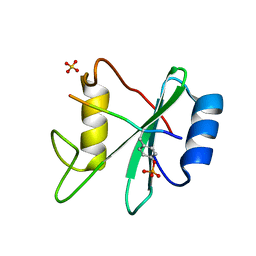 | | Structure of SHP2 N-SH2 domain in complex with RLNpYAQLWHR peptide | | Descriptor: | RLNpYAQLWHR peptide, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Y, Zhang, J, Yuan, C, Hard, R.L, Park, I.H, Li, C, Bell, C.E, Pei, D. | | Deposit date: | 2011-08-29 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Simultaneous binding of two peptidyl ligands by a SRC homology 2 domain.
Biochemistry, 50, 2011
|
|
4CN4
 
 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant with 2-deoxy- 2-fluoro-beta-maltosyl modification | | Descriptor: | ALPHA-1,4-GLUCAN:MALTOSE-1-PHOSPHATE MALTOSYLTRANSFERASE 1, alpha-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-beta-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rashid, A.M, Saalbach, G, Tang, M, Tuukanen, A, Svergun, D.I, Withers, S.G, Lawson, D.M, Bornemann, S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight Into How Streptomyces Coelicolor Maltosyl Transferase Glge Binds Alpha-Maltose 1-Phosphate and Forms a Maltosyl-Enzyme Intermediate.
Biochemistry, 53, 2014
|
|
5SWD
 
 | | Structure of the adenine riboswitch aptamer domain in an intermediate-bound state | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, ... | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
6CUT
 
 | | Engineered Holo TrpB from Pyrococcus furiosus, PfTrpB7E6 with (2S,3S)-isopropylserine bound as the external aldimine | | Descriptor: | (2S,3S)-3-hydroxy-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-methylpentanoic acid (non-preferred name), SODIUM ION, Tryptophan synthase beta chain 1 | | Authors: | Boville, C.E, Scheele, R.A, Buller, A.R, Arnold, F.H. | | Deposit date: | 2018-03-26 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Engineered Biosynthesis of beta-Alkyl Tryptophan Analogues.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5SWE
 
 | | Ligand-bound structure of adenine riboswitch aptamer domain converted in crystal from its ligand-free state using ligand mixing serial femtosecond crystallography | | Descriptor: | ADENINE, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
4CN6
 
 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant with maltose bound | | Descriptor: | ALPHA-1,4-GLUCAN:MALTOSE-1-PHOSPHATE MALTOSYLTRANSFERASE 1, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rashid, A.M, Saalbach, G, Tang, M, Tuukanen, A, Svergun, D.I, Withers, S.G, Lawson, D.M, Bornemann, S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Insight Into How Streptomyces Coelicolor Maltosyl Transferase Glge Binds Alpha-Maltose 1-Phosphate and Forms a Maltosyl-Enzyme Intermediate.
Biochemistry, 53, 2014
|
|
4CVJ
 
 | | Neutron Structure of Compound I intermediate of Cytochrome c Peroxidase - Deuterium exchanged 100 K | | Descriptor: | CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Casadei, C.M, Gumiero, A, Blakeley, M.P, Ostermann, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (2.182 Å), X-RAY DIFFRACTION | | Cite: | Neutron Cryo-Crystallography Captures the Protonation State of Ferryl Heme in a Peroxidase
Science, 345, 2014
|
|
4CVI
 
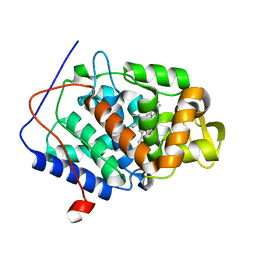 | | Neutron Structure of Ferric Cytochrome c Peroxidase - Deuterium exchanged at room temperature | | Descriptor: | CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Casadei, C.M, Gumiero, A, Blakeley, M.P, Ostermann, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (2.1 Å), X-RAY DIFFRACTION | | Cite: | Neutron Cryo-Crystallography Captures the Protonation State of Ferryl Heme in a Peroxidase
Science, 345, 2014
|
|
6D1O
 
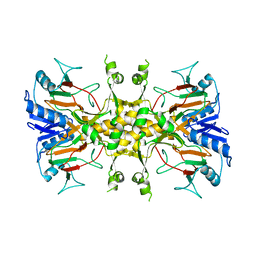 | | FT_5 dioxygenase apoenzyme | | Descriptor: | (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase | | Authors: | Rydel, T.J, Halls, C.E. | | Deposit date: | 2018-04-12 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
6BZ6
 
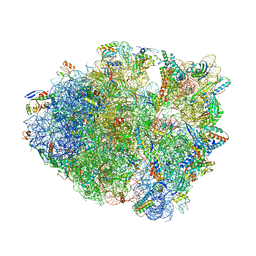 | | Thermus thermophilus 70S complex containing 16S G347U ram mutation and empty A site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2017-12-22 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding.
Nucleic Acids Res., 47, 2019
|
|
5U50
 
 | |
4CKK
 
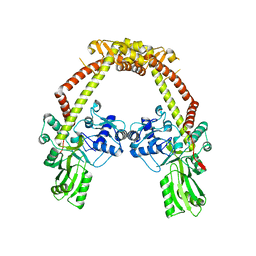 | | Apo structure of 55 kDa N-terminal domain of E. coli DNA gyrase A subunit | | Descriptor: | DNA GYRASE SUBUNIT A | | Authors: | Hearnshaw, S.J, Edwards, M.J, Stevenson, C.E.M, Lawson, D.M, Maxwell, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A New Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8 Bound to DNA Gyrase Gives Fresh Insight Into the Mechanism of Inhibition.
J.Mol.Biol., 426, 2014
|
|
3R5W
 
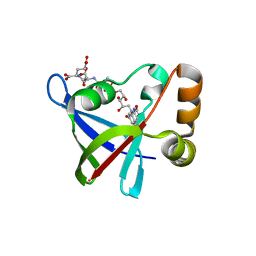 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5P
 
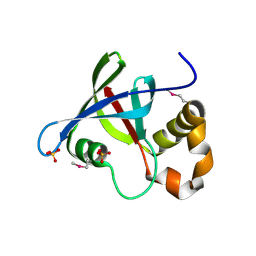 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | Deazaflavin-dependent nitroreductase, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R98
 
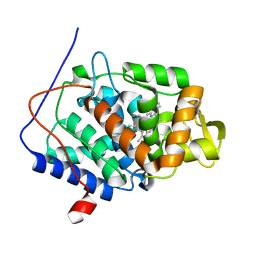 | | Joint Neutron and X-ray structure of Cytochrome c peroxidase | | Descriptor: | Cytochrome c peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Blakeley, M.P, Fisher, S.J, Gumiero, A, Moody, P.C.E, Raven, E.L. | | Deposit date: | 2011-03-25 | | Release date: | 2012-04-04 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (2.4 Å), X-RAY DIFFRACTION | | Cite: | Hydrogen bonds in heme peroxidases: a combined X-ray and neutron study of cytochrome c peroxidase
To be Published
|
|
3R5Y
 
 | | Structure of a Deazaflavin-dependent nitroreductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3RLL
 
 | | Crystal structure of the T877A androgen receptor ligand binding domain in complex with (S)-N-(4-Cyano-3-(trifluoromethyl)phenyl)-3-(4-cyanonaphthalen-1-yloxy)-2-hydroxy-2-methylpropanamide | | Descriptor: | (2S)-3-[(4-cyanonaphthalen-1-yl)oxy]-N-[4-cyano-3-(trifluoromethyl)phenyl]-2-hydroxy-2-methylpropanamide, Androgen receptor | | Authors: | Bohl, C.E, Duke, C.B, Jones, A, Dalton, J.T, Miller, D.D. | | Deposit date: | 2011-04-19 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unexpected binding orientation of bulky-B-ring anti-androgens and implications for future drug targets.
J.Med.Chem., 54, 2011
|
|
7QT2
 
 | | Antibody FenAb208 - fentanyl complex | | Descriptor: | Antibody heavy chain, Antibody light chain, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2022-01-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A trypanosome-derived immunotherapeutics platform elicits potent high-affinity antibodies, negating the effects of the synthetic opioid fentanyl.
Cell Rep, 42, 2023
|
|
