4GSO
 
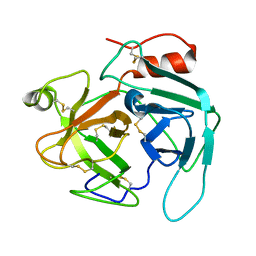 | | structure of Jararacussin-I | | Descriptor: | Thrombin-like enzyme BjussuSP-1 | | Authors: | Ullah, A, Souza, T.C.A.B, Zanphorlin, L.M, Mariutti, R, Sanata, S.V, Murakami, M.T, Arni, R.K. | | Deposit date: | 2012-08-28 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Jararacussin-I: The highly negatively charged catalytic interface contributes to macromolecular selectivity in snake venom thrombin-like enzymes.
Protein Sci., 22, 2013
|
|
4AQW
 
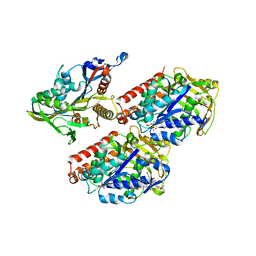 | | Model of human kinesin-5 motor domain (1II6, 3HQD) and mammalian tubulin heterodimer (1JFF) docked into the 9.5-angstrom cryo-EM map of microtubule-bound kinesin-5 motor domain in the rigor state. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-LIKE PROTEIN KIF11, ... | | Authors: | Goulet, A, Behnke-Parks, W.M, Sindelar, C.V, Rosenfeld, S.S, Moores, C.A. | | Deposit date: | 2012-04-19 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | The Structural Basis of Force Generation by the Mitotic Motor Kinesin-5.
J.Biol.Chem., 287, 2012
|
|
4H55
 
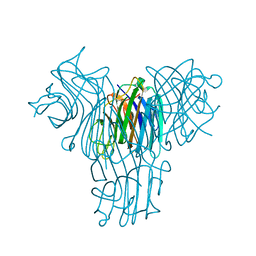 | | Crystal structure of Canavalia brasiliensis seed lectin (ConBr) in complex with beta-d-ribofuranose | | Descriptor: | CALCIUM ION, Concanavalin-Br, D-ALPHA-AMINOBUTYRIC ACID, ... | | Authors: | Salviano, E, Rocha, B.A.M, Cavada, B.S, Delatorre, P, Santi-Gadelha, T, Gadelha, C.A.A, Silva-Filho, J.C, Nobrega, R.B, Farias, D.L. | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of Canavalia brasiliensis seed lectin (ConBr) in complex with beta-d-ribofuranose
TO BE PUBLISHED
|
|
1E91
 
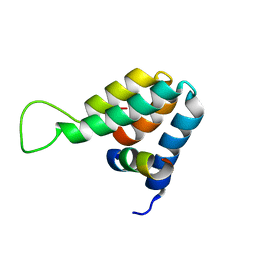 | | Structure of the complex of the Mad1-Sin3B interaction domains | | Descriptor: | MAD PROTEIN (MAX DIMERIZER), PAIRED AMPHIPATHIC HELIX PROTEIN SIN3B | | Authors: | Spronk, C.A.E.M, Tessari, M, Kaan, A.M, Jansen, J.F.A, Vermeulen, M, Stunnenberg, H.G, Vuister, G.W. | | Deposit date: | 2000-10-04 | | Release date: | 2000-11-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The MAD1-Sin3B Interaction Involves a Novel Helical Fold
Nat.Struct.Biol., 7, 2000
|
|
1H95
 
 | | Solution structure of the single-stranded DNA-binding Cold Shock Domain (CSD) of human Y-box protein 1 (YB1) determined by NMR (10 lowest energy structures) | | Descriptor: | Y-BOX BINDING PROTEIN | | Authors: | Kloks, C.P.A.M, Spronk, C.A.E.M, Hoffmann, A, Vuister, G.W, Grzesiek, S, Hilbers, C.W. | | Deposit date: | 2001-02-23 | | Release date: | 2002-02-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure and DNA-Binding Properties of the Cold-Shock Domain of the Human Y-Box Protein Yb-1.
J.Mol.Biol., 316, 2002
|
|
4DCQ
 
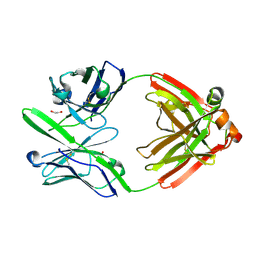 | | Crystal Structure of the Fab Fragment of 3B5H10, an Antibody-Specific for Extended Polyglutamine Repeats (orthorhombic form) | | Descriptor: | 1,2-ETHANEDIOL, 3B5H10 FAB Heavy Chain, 3B5H10 FAB Light Chain | | Authors: | Peters-Libeu, C.A, Tran, T, Finkbeiner, S, Weisgraber, K. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
1VJH
 
 | | Crystal structure of gene product of At1g24000 from Arabidopsis thaliana | | Descriptor: | Bet v I allergen family | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1H, 15N and 13C resonance assignments of the putative Bet v 1 family protein At1g24000.1 from Arabidopsis thaliana.
J.Biomol.Nmr, 32, 2005
|
|
1VM0
 
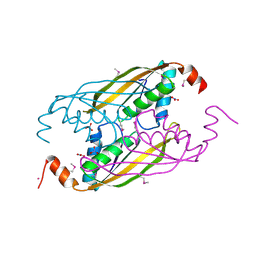 | | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT2G34160 | | Descriptor: | NITRATE ION, POTASSIUM ION, unknown protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-08-24 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT2G34160
To be published
|
|
1VKP
 
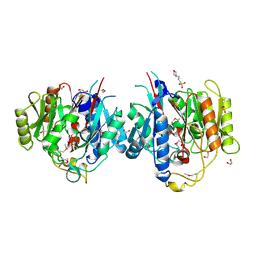 | | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT5G08170, AGMATINE IMINOHYDROLASE | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, AGMATINE IMINOHYDROLASE, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-06-15 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g08170
To be published
|
|
4I30
 
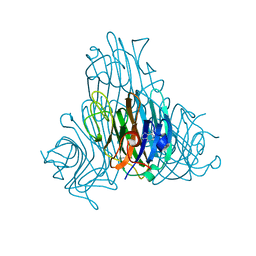 | | Crystal structure of Canavalia maritima seeds lectin (ConM) co-crystalized with gamma-aminobutyric acid (GABA) and soaked with adenine | | Descriptor: | ADENINE, CALCIUM ION, Concanavalin-A, ... | | Authors: | Delatorre, P, Silva-Filho, J.C, Nobrega, R.B, Gadelha, C.A.A, Cavada, B.S, Rocha, B.A.M, Santi-Gadelha, T, Teixeira, C.S. | | Deposit date: | 2012-11-23 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of Canavalia maritima seeds lectin (ConM) co-crystalized with gamma-aminobutyric acid (GABA) and soaked with adenine
To be Published
|
|
3D89
 
 | | Crystal Structure of a Soluble Rieske Ferredoxin from Mus musculus | | Descriptor: | 1,2-ETHANEDIOL, FE2/S2 (INORGANIC) CLUSTER, Rieske domain-containing protein | | Authors: | Levin, E.J, McCoy, J.G, Elsen, N.L, Seder, K.D, Bingman, C.A, Wesenberg, G.E, Fox, B.G, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-05-22 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.071 Å) | | Cite: | X-ray structure of a soluble Rieske-type ferredoxin from Mus musculus.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3PAZ
 
 | | REDUCED NATIVE PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-20 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
3EMM
 
 | | X-ray structure of protein from Arabidopsis thaliana AT1G79260 with Bound Heme | | Descriptor: | 1,2-ETHANEDIOL, PROTOPORPHYRIN IX CONTAINING FE, Uncharacterized protein At1g79260 | | Authors: | Bianchetti, C.M, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-09-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.358 Å) | | Cite: | The structure and NO binding properties of the nitrophorin-like heme-binding protein from Arabidopsis thaliana gene locus At1g79260.1.
Proteins, 78, 2010
|
|
5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
3DKV
 
 | | Crystal structure of adenylate kinase variant AKlse1 | | Descriptor: | 1,2-ETHANEDIOL, Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Bannen, R.M, Bianchetti, C.M, Bingman, C.A, Bitto, E.B. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of adenylate kinase variant AKlse1.
To be Published
|
|
5KSQ
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa | | Descriptor: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KSS
 
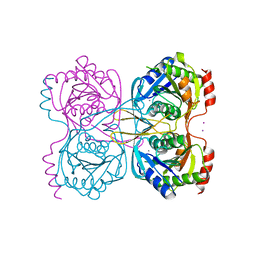 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-Ds (Dimer Smaller) | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, A.M.S, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KST
 
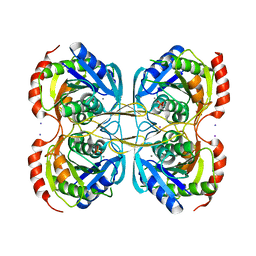 | | Stationary phase Survival protein E (SurE) from Xylella fastidiosa- XfSurE-TSAmp (Tetramer Smaller - crystallization with 3'AMP). | | Descriptor: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A.S, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.759 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
3IAU
 
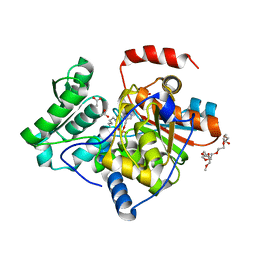 | | The structure of the processed form of threonine deaminase isoform 2 from Solanum lycopersicum | | Descriptor: | ACETATE ION, POLYETHYLENE GLYCOL (N=34), SULFATE ION, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-07-14 | | Release date: | 2009-07-28 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Adaptive evolution of threonine deaminase in plant defense against insect herbivores.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3F61
 
 | |
3F69
 
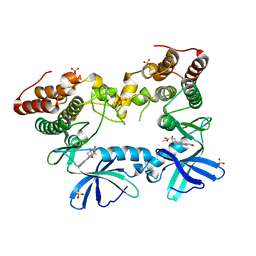 | | Crystal structure of the Mycobacterium tuberculosis PknB mutant kinase domain in complex with KT5720 | | Descriptor: | SULFATE ION, Serine/threonine-protein kinase pknB, hexyl (5S,6R,8R)-6-hydroxy-5-methyl-13-oxo-5,6,7,8-tetrahydro-13H-5,8-epoxy-4b,8a,14-triazadibenzo[b,h]cycloocta[1,2,3,4-jkl]c yclopenta[e]-as-indacene-6-carboxylate | | Authors: | Alber, T, Mieczkowski, C.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Auto-activation mechanism of the Mycobacterium tuberculosis PknB receptor Ser/Thr kinase.
Embo J., 27, 2008
|
|
5LTU
 
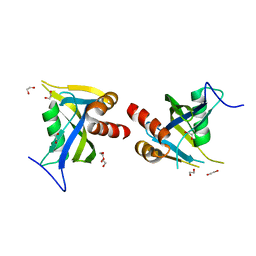 | | Crystal Structure of NUDT4A- Diphosphoinositol polyphosphate phosphohydrolase 2 | | Descriptor: | 1,2-ETHANEDIOL, Diphosphoinositol polyphosphate phosphohydrolase 2 | | Authors: | Srikannathasan, V, Nunez, C.A, Tallant, C, Siejka, P, Mathea, S, Newman, J, Strain-Damerell, C, Elkins, J.M, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structure of Human NUDT4A- Diphosphoinositol polyphosphate phosphohydrolase 2
To Be Published
|
|
5LQP
 
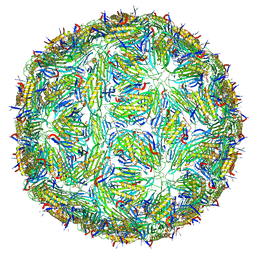 | |
2AMY
 
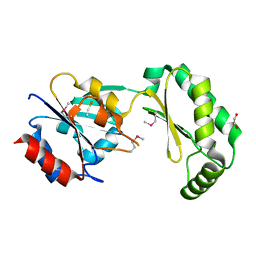 | | X-Ray Structure of Human Phosphomannomutase 2 (PMM2) | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, Phosphomannomutase 2 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-08-10 | | Release date: | 2005-08-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | X-Ray Structure of Human Phosphomannomutase 2 (PMM2)
To be Published
|
|
3RS6
 
 | | Crystal structure Dioclea virgata lectin in complexed with X-mannose | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Lectin alpha chain, ... | | Authors: | Gadelha, C.A.A, Santi-Gadelha, T, Nagano, C.S, Bezerra, E.H.S, Bezerra, M.J.B, Alencar, K.L, Silva-Filho, J.C. | | Deposit date: | 2011-05-02 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Dioclea virgata lectin: Relations between carbohydrate binding site and nitric oxide production.
Biochimie, 94, 2012
|
|
