1F5C
 
 | | CRYSTAL STRUCTURE OF F25H FERREDOXIN 1 MUTANT FROM AZOTOBACTER VINELANDII AT 1.75 ANGSTROM RESOLUTION | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN 1, IRON/SULFUR CLUSTER, ... | | Authors: | Chen, K, Bonagura, C.A, Tilley, G.J, Jung, Y.S, Armstrong, F.A, Stout, C.D, Burgess, B.K. | | Deposit date: | 2000-06-13 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of ferredoxin variants exhibiting large changes in [Fe-S] reduction potential.
Nat.Struct.Biol., 9, 2002
|
|
1F02
 
 | | CRYSTAL STRUCTURE OF C-TERMINAL 282-RESIDUE FRAGMENT OF INTIMIN IN COMPLEX WITH TRANSLOCATED INTIMIN RECEPTOR (TIR) INTIMIN-BINDING DOMAIN | | Descriptor: | INTIMIN, TRANSLOCATED INTIMIN RECEPTOR | | Authors: | Luo, Y, Frey, E.A, Pfuetzner, R.A, Creagh, A.L, Knoechel, D.G, Haynes, C.A, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2000-05-14 | | Release date: | 2000-07-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex.
Nature, 405, 2000
|
|
1F5B
 
 | | CRYSTAL STRUCTURE OF F2H FERREDOXIN 1 MUTANT FROM AZOTOBACTER VINELANDII AT 1.75 ANGSTROM RESOLUTION | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN 1, IRON/SULFUR CLUSTER | | Authors: | Chen, K, Bonagura, C.A, Tilley, G.J, Jung, Y.S, Armstrong, F.A, Stout, C.D, Burgess, B.K. | | Deposit date: | 2000-06-13 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structures of ferredoxin variants exhibiting large changes in [Fe-S] reduction potential.
Nat.Struct.Biol., 9, 2002
|
|
1F1C
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C549 | | Descriptor: | CYTOCHROME C549, HEME C | | Authors: | Kerfeld, C.A, Sawaya, M.R, Yeates, T.O, Krogmann, D.W. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
1DUV
 
 | | CRYSTAL STRUCTURE OF E. COLI ORNITHINE TRANSCARBAMOYLASE COMPLEXED WITH NDELTA-L-ORNITHINE-DIAMINOPHOSPHINYL-N-SULPHONIC ACID (PSORN) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NDELTA-(N'-SULPHODIAMINOPHOSPHINYL)-L-ORNITHINE, ORNITHINE TRANSCARBAMOYLASE | | Authors: | Langley, D.B, Templeton, M.D, Fields, B.A, Mitchell, R.E, Collyer, C.A. | | Deposit date: | 2000-01-18 | | Release date: | 2000-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of inactivation of ornithine transcarbamoylase by Ndelta -(N'-Sulfodiaminophosphinyl)-L-ornithine, a true transition state analogue? Crystal structure and implications for catalytic mechanism.
J.Biol.Chem., 275, 2000
|
|
8SF4
 
 | |
1DTG
 
 | | HUMAN TRANSFERRIN N-LOBE MUTANT H249E | | Descriptor: | CARBONATE ION, FE (III) ION, TRANSFERRIN | | Authors: | MacGillivray, R.T, Bewley, M.C, Smith, C.A, He, Q.Y, Mason, A.B. | | Deposit date: | 2000-01-12 | | Release date: | 2000-01-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mutation of the iron ligand His 249 to Glu in the N-lobe of human transferrin abolishes the dilysine "trigger" but does not significantly affect iron release.
Biochemistry, 39, 2000
|
|
2LUZ
 
 | | Solution NMR Structure of CalU16 from Micromonospora echinospora, Northeast Structural Genomics Consortium (NESG) Target MiR12 | | Descriptor: | CalU16 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Pederson, K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Wrobel, R.L, Bingman, C.A, Singh, S, Thorson, J.S, Prestegard, J.H, Montelione, G.T, Phillips Jr, G.N, Kennedy, M.A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-22 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Acs Chem.Biol., 9, 2014
|
|
1H0H
 
 | | Tungsten containing Formate Dehydrogenase from Desulfovibrio Gigas | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Raaijmakers, H.C.A. | | Deposit date: | 2002-06-19 | | Release date: | 2003-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gene Sequence and the 1.8 A Crystal Structure of the Tungsten-Containing Formate Dehydrogenase from Desulfovibrio Gigas
Structure, 10, 2002
|
|
2XMY
 
 | | Discovery and Characterisation of 2-Anilino-4-(thiazol-5-yl) pyrimidine Transcriptional CDK Inhibitors as Anticancer Agents | | Descriptor: | 4-[4-(3,4-DIMETHYL-2-OXO-2,3-DIHYDRO-THIAZOL-5-YL)-PYRIMIDIN-2-YLAMINO]-N-(2-METHOXY-ETHYL)-BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wang, S, Griffiths, G, Midgley, C.A, Barnett, A.L, Cooper, M, Grabarek, J, Ingram, L, Jackson, W, Kontopidis, G, McClue, S.J, McInnes, C, McLachlan, J, Meades, C, Mezna, M, Stuart, I, Thomas, M.P, Zheleva, D.I, Lane, D.P, Jackson, R.C, Glover, D.M, Blake, D.G, Fischer, P.M. | | Deposit date: | 2010-07-29 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Characterisation of 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Transcriptional Cdk Inhibitors as Anticancer Agents
Chem.Biol., 17, 2010
|
|
2M5K
 
 | | Atomic-resolution structure of a doublet cross-beta amyloid fibril | | Descriptor: | Transthyretin | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-12-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.7 Å), SOLID-STATE NMR | | Cite: | Atomic structure and hierarchical assembly of a cross-beta amyloid fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2M5M
 
 | | Atomic-resolution structure of a triplet cross-beta amyloid fibril | | Descriptor: | Transthyretin | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-12-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.2 Å), SOLID-STATE NMR | | Cite: | Atomic structure and hierarchical assembly of a cross-beta amyloid fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8FAJ
 
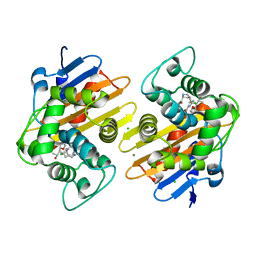 | | OXA-48-NA-1-157 inhibitor complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Stewart, N.K, Toth, M, Vakulenko, S.B. | | Deposit date: | 2022-11-28 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Exhibits Potent Activity against Klebsiella spp. Isolates Producing OXA-48-Type Carbapenemases.
Acs Infect Dis., 9, 2023
|
|
2XRP
 
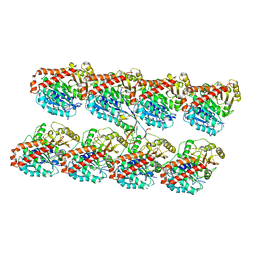 | | Human Doublecortin N-DC Repeat (1MJD) and Mammalian Tubulin (1JFF and 3HKE) Docked into the 8-Angstrom Cryo-EM Map of Doublecortin- Stabilised Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, NEURONAL MIGRATION PROTEIN DOUBLECORTIN, ... | | Authors: | Fourniol, F.J, Sindelar, C.V, Amigues, B, Clare, D.K, Thomas, G, Perderiset, M, Francis, F, Houdusse, A, Moores, C.A. | | Deposit date: | 2010-09-18 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Template-Free 13-Protofilament Microtubule-Map Assembly Visualized at 8 A Resolution.
J.Cell Biol., 191, 2010
|
|
1EXF
 
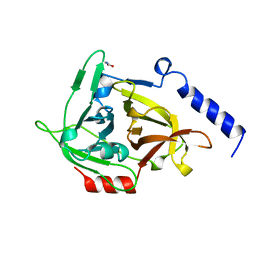 | | EXFOLIATIVE TOXIN A | | Descriptor: | EXFOLIATVE TOXIN A, GLYCINE | | Authors: | Vath, G.M, Earhart, C.A, Rago, J.V, Kim, M.H, Bohach, G.A, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-10-22 | | Release date: | 1998-02-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the superantigen exfoliative toxin A suggests a novel regulation as a serine protease.
Biochemistry, 36, 1997
|
|
2N1L
 
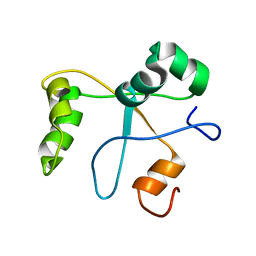 | | Solution structure of the BCOR PUFD | | Descriptor: | BCL-6 corepressor | | Authors: | Wong, S.J, Gearhart, M.D, Ha, D.J, Corcoran, C.M, Diaz, V, Taylor, A.B, Schirf, V, Ilangovan, U, Hinck, A.P, Demeler, B, Hart, J, Bardwell, V.J, Kim, C.A. | | Deposit date: | 2015-04-06 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the hierarchical assembly of the core of PRC1.1
To be Published
|
|
2MZ9
 
 | | Solution structure of oxidized triheme cytochrome PpcA from Geobacter sulfurreducens | | Descriptor: | HEME C, PpcA | | Authors: | Morgado, L, Bruix, M, Pokkuluri, R, Salgueiro, C.A, Turner, D.L. | | Deposit date: | 2015-02-08 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Redox- and pH-linked conformational changes in triheme cytochrome PpcA from Geobacter sulfurreducens.
Biochem. J., 474, 2017
|
|
1F1F
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM ARTHROSPIRA MAXIMA | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Serag, A.A, Sawaya, M.R, Krogmann, D.W, Yeates, T.O. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
2Y1S
 
 | | Microvirin lectin | | Descriptor: | Mannan-binding lectin | | Authors: | Bewley, C.A, Hussan, S. | | Deposit date: | 2010-12-10 | | Release date: | 2011-04-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the monovalent lectin microvirin in complex with Man(alpha)(1-2)Man provides a basis for anti-HIV activity with low toxicity.
J. Biol. Chem., 286, 2011
|
|
8FL5
 
 | |
1E3J
 
 | | Ketose reductase (sorbitol dehydrogenase) from silverleaf whitefly | | Descriptor: | BORIC ACID, NADP(H)-DEPENDENT KETOSE REDUCTASE, PHOSPHATE ION, ... | | Authors: | Banfield, M.J, Salvucci, M.E, Baker, E.N, Smith, C.A. | | Deposit date: | 2000-06-19 | | Release date: | 2001-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Nadp(H)-Dependent Ketose Reductase from Besimia Argentifolii at 2.3 Angstrom Resolution
J.Mol.Biol., 306, 2001
|
|
2Y5A
 
 | | Cytochrome c peroxidase (CCP) W191G bound to 3-aminopyridine | | Descriptor: | 3-AMINOPYRIDINE, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Cappel, D, Wahlstrom, R, Brenk, R, Sotriffer, C.A. | | Deposit date: | 2011-01-12 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Probing the Dynamic Nature of Water Molecules and Their Influences on Ligand Binding in a Model Binding Site.
J.Chem.Inf.Model, 51, 2011
|
|
8TNS
 
 | |
1FCK
 
 | | STRUCTURE OF DICERIC HUMAN LACTOFERRIN | | Descriptor: | CARBONATE ION, CERIUM (III) ION, LACTOFERRIN | | Authors: | Baker, H.M, Baker, C.J, Smith, C.A, Baker, E.N. | | Deposit date: | 2000-07-18 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Metal substitution in transferrins: specific binding of cerium(IV) revealed by the crystal structure of cerium-substituted human lactoferrin.
J.Biol.Inorg.Chem., 5, 2000
|
|
1FH1
 
 | |
