8AQ9
 
 | |
4IZQ
 
 | |
4L7Z
 
 | | Crystal Structure of Chloroflexus aurantiacus malyl-CoA lyase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase | | Authors: | Zarzycki, J, Kerfeld, C.A. | | Deposit date: | 2013-06-14 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | The crystal structures of the tri-functional Chloroflexus aurantiacus and bi-functional Rhodobacter sphaeroides malyl-CoA lyases and comparison with CitE-like superfamily enzymes and malate synthases.
Bmc Struct.Biol., 13, 2013
|
|
4KX3
 
 | |
6Q2J
 
 | | Cryo-EM structure of extracellular dimeric complex of RET/GFRAL/GDF15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-like, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
4J3Y
 
 | | Crystal structure of XIAP-BIR2 domain | | Descriptor: | E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2K14
 
 | | Solution structure of the soluble domain of the NfeD protein YuaF from Bacillus subtilis | | Descriptor: | YuaF protein | | Authors: | Walker, C.A, Hinderhofer, M, Witte, D.J, Boos, W, Moller, H.M. | | Deposit date: | 2008-02-20 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the soluble domain of the NfeD protein YuaF from Bacillus subtilis.
J.Biomol.Nmr, 42, 2008
|
|
1JDR
 
 | | Crystal Structure of a Proximal Domain Potassium Binding Variant of Cytochrome c Peroxidase | | Descriptor: | Cytochrome c Peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bonagura, C.A, Sundaramoorthy, M, Bhaskar, B, Poulos, T.L. | | Deposit date: | 2001-06-14 | | Release date: | 2001-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The effects of an engineered cation site on the structure, activity, and EPR properties of cytochrome c peroxidase.
Biochemistry, 38, 1999
|
|
6PYS
 
 | | Human PI3Kalpha in complex with Compound 2-10 ((3S)-3-benzyl-3-methyl-5-[5-(2-methylpyrimidin-5-yl)pyrazolo[1,5-a]pyrimidin-3-yl]-1,3-dihydro-2H-indol-2-one) | | Descriptor: | (3S)-3-benzyl-3-methyl-5-[5-(2-methylpyrimidin-5-yl)pyrazolo[1,5-a]pyrimidin-3-yl]-1,3-dihydro-2H-indol-2-one, GLYCEROL, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Lesburg, C.A, Augustin, M.A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Design of selective PI3K delta inhibitors using an iterative scaffold-hopping workflow.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6Q1I
 
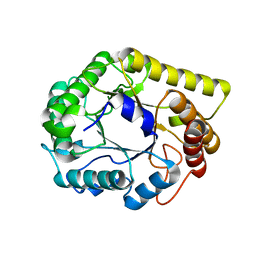 | |
4L7I
 
 | | Crystal structure of S-Adenosylmethionine synthase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-13 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
2F2G
 
 | | X-Ray Structure of Gene Product From Arabidopsis Thaliana AT3G16990 | | Descriptor: | 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, SEED MATURATION PROTEIN PM36 HOMOLOG, SULFATE ION | | Authors: | Wesenberg, G.W, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-11-16 | | Release date: | 2005-12-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of gene locus At3g16990 from Arabidopsis thaliana
Proteins, 57, 2004
|
|
6Q2S
 
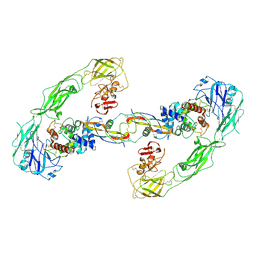 | | Cryo-EM structure of RET/GFRa3/ARTN extracellular complex. The 3D refinement was applied with C2 symmetry. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-3, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
4ITC
 
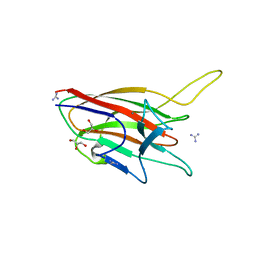 | | Crystal Structure Analysis of the K1 Cleaved Adhesin domain of Lys-gingipain (Kgp) from Porphyromonas gingivalis W83 | | Descriptor: | CALCIUM ION, GLYCEROL, GUANIDINE, ... | | Authors: | Ganuelas, L.A, Li, N, Hunter, N, Collyer, C.A. | | Deposit date: | 2013-01-18 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The lysine gingipain adhesin domains from Porphyromonas gingivalis interact with erythrocytes and albumin: Structures correlate to function.
Eur J Microbiol Immunol (Bp), 3, 2013
|
|
1CPT
 
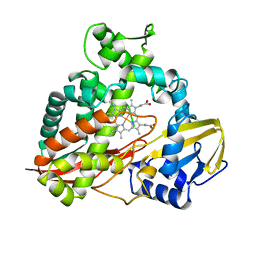 | | CRYSTAL STRUCTURE AND REFINEMENT OF CYTOCHROME P450-TERP AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME P450-TERP, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hasemann, C.A, Ravichandran, K.G, Peterson, J.A, Deisenhofer, J. | | Deposit date: | 1993-11-23 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and refinement of cytochrome P450terp at 2.3 A resolution.
J.Mol.Biol., 236, 1994
|
|
1JBV
 
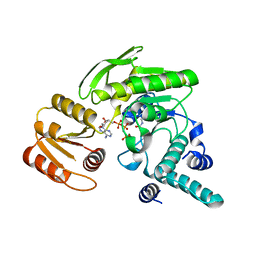 | | FPGS-AMPPCP complex | | Descriptor: | FOLYLPOLYGLUTAMATE SYNTHASE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Sun, X, Cross, J.A, Bognar, A.L, Baker, E.N, Smith, C.A. | | Deposit date: | 2001-06-06 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Folate-binding triggers the activation of folylpolyglutamate synthetase.
J.Mol.Biol., 310, 2001
|
|
2F8K
 
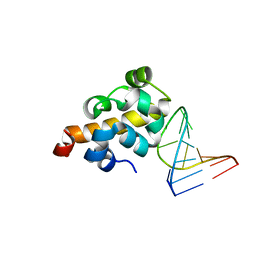 | | Sequence specific recognition of RNA hairpins by the SAM domain of Vts1 | | Descriptor: | 5'-R(*UP*AP*AP*UP*CP*UP*UP*UP*GP*AP*CP*AP*GP*AP*UP*U)-3', Protein VTS1 | | Authors: | Aviv, T, Lin, Z, Ben-Ari, G, Smibert, C.A, Sicheri, F. | | Deposit date: | 2005-12-02 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1JIL
 
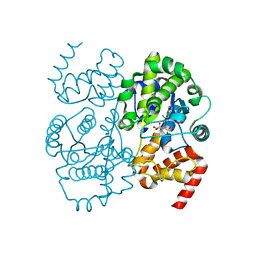 | | Crystal structure of S. aureus TyrRS in complex with SB284485 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]- (3,4,5-TRIHYDROXY-6-METHYL-TETRAHYDRO-PYRAN-2-YL)- ACETIC ACID, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
7ZLV
 
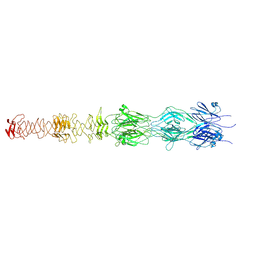 | | Tail tip of siphophage T5 : central fibre protein pb4 | | Descriptor: | Probable central straight fiber | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2022-04-15 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
4ISZ
 
 | | RNA ligase RtcB in complex with GTP alphaS and Mn(II) | | Descriptor: | GUANOSINE-5'-RP-ALPHA-THIO-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-01-17 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structures of the Noncanonical RNA Ligase RtcB Reveal the Mechanism of Histidine Guanylylation.
Biochemistry, 52, 2013
|
|
6PZ7
 
 | | GH5-4 broad specificity endoglucanase from Clostridium acetobutylicum | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Endoglucanase family 5 | | Authors: | Bianchetti, C.M, Bingman, C.A, Fox, B.G. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
4GIP
 
 | | Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 (PIV5) fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F1, Fusion glycoprotein F2 | | Authors: | Welch, B.D, Liu, Y, Kors, C.A, Leser, G.P, Jardetzky, T.S, Lamb, R.A. | | Deposit date: | 2012-08-08 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6Q2O
 
 | | Cryo-EM structure of RET/GFRa2/NRTN extracellular complex. The 3D refinement was applied with C2 symmetry. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-2, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
2EZM
 
 | |
4MZ4
 
 | | Discovery of an Irreversible HCV NS5B Polymerase Inhibitor | | Descriptor: | 1-[(2-chloroquinolin-3-yl)methyl]-6-fluoro-5-methyl-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Zeng, Q, Anilkumar, G.N, Rosenblum, S.B, Huang, H.-C, Lesburg, C.A, Jiang, Y, Selyutin, O, Chan, T.-Y, Bennett, F, Chen, K.X, Venkatraman, S, Sannigrahi, M, Velazquez, F, Duca, J.S, Gavalas, S, Huang, Y, Pu, H, Wang, L, Pinto, P, Vibulbhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Li, C, Hesk, D, Gesell, J, Sorota, S, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2013-09-29 | | Release date: | 2013-12-11 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of an irreversible HCV NS5B polymerase inhibitor.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
