6TA3
 
 | | Human kinesin-5 motor domain in the GSK-1 state bound to microtubules (Conformation 1) | | Descriptor: | 6-[4-(trifluoromethyl)phenyl]-3,4-dihydro-1~{H}-quinolin-2-one, Kinesin-like protein KIF11, MAGNESIUM ION, ... | | Authors: | Pena, A, Sweeney, A, Cook, A.D, Moores, C.A, Topf, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Microtubule-Trapped Human Kinesin-5 and Its Mechanism of Inhibition Revealed Using Cryoelectron Microscopy.
Structure, 28, 2020
|
|
2WLY
 
 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitotrio-thiazoline. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-29 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chitinase inhibition by chitobiose and chitotriose thiazolines.
Angew. Chem. Int. Ed. Engl., 49, 2010
|
|
1GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
1RWJ
 
 | | c7-type three-heme cytochrome domain | | Descriptor: | Cytochrome c family protein, HEME C | | Authors: | Pokkuluri, P.R, Londer, Y.Y, Duke, N.E.C, Erickson, J, Pessanha, M, Salgueiro, C.A, Schiffer, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-08-03 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a novel c7-type three-heme cytochrome domain from a multidomain cytochrome c polymer.
Protein Sci., 13, 2004
|
|
5CG2
 
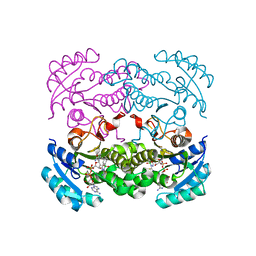 | | Crystal structure of E. coli FabI bound to the thiocarbamoylated benzodiazaborine inhibitor 35b. | | Descriptor: | 1-hydroxy-2,3,1-benzodiazaborinine-2(1H)-carbothioamide, Enoyl-[acyl-carrier-protein] reductase [NADH] FabI, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jordan, C.A, Vey, J.L. | | Deposit date: | 2015-07-09 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystallographic insights into the structure-activity relationships of diazaborine enoyl-ACP reductase inhibitors.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5CFZ
 
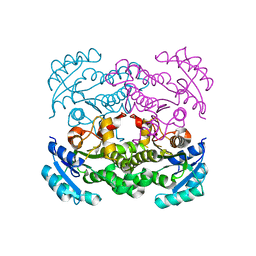 | | Crystal structure of E. coli FabI in apo form | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH] FabI | | Authors: | Jordan, C.A, Vey, J.L. | | Deposit date: | 2015-07-08 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystallographic insights into the structure-activity relationships of diazaborine enoyl-ACP reductase inhibitors.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2V3G
 
 | | Structure of a family 26 lichenase in complex with noeuromycin | | Descriptor: | (2R,3S,4R,5R)-5-(HYDROXYMETHYL)PIPERIDINE-2,3,4-TRIOL, ENDOGLUCANASE H, beta-D-glucopyranose | | Authors: | Meloncelli, P.J, Gloster, T.M, Money, V.A, Tarling, C.A, Davies, G.J, Withers, S.G, Stick, R.V. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | D-Glucosylated Derivatives of Isofagomine and Noeuromycin and Their Potential as Inhibitors of Beta-Glycoside Hydrolases
To be Published
|
|
5CUO
 
 | |
5CTN
 
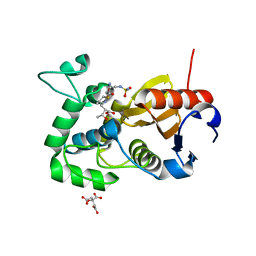 | | Structure of BPu1 beta-lactamase | | Descriptor: | (2~{S},3~{R})-3-methyl-2-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase, CITRATE ANION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-07-24 | | Release date: | 2015-11-25 | | Last modified: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Class D beta-lactamases do exist in Gram-positive bacteria.
Nat.Chem.Biol., 12, 2016
|
|
5CUP
 
 | |
5CV3
 
 | | C. remanei PGL-1 Dimerization Domain - Hg | | Descriptor: | ETHYL MERCURY ION, Putative uncharacterized protein | | Authors: | Aoki, S.T, Bingman, C.A, Wickens, M, Kimble, J.E. | | Deposit date: | 2015-07-25 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.17014766 Å) | | Cite: | PGL germ granule assembly protein is a base-specific, single-stranded RNase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5WO2
 
 | | Chaperone Spy bound to Casein Fragment (Casein un-modeled) | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Periplasmic chaperone Spy, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5A2S
 
 | | Potent, selective and CNS-penetrant tetrasubstituted cyclopropane class IIa histone deacetylase (HDAC) inhibitors | | Descriptor: | (1S,2S,3S)-1-fluoranyl-2-[4-(5-fluoranylpyrimidin-2-yl)phenyl]-N-oxidanyl-3-phenyl-cyclopropane-1-carboxamide, HISTONE DEACETYLASE 4, SODIUM ION, ... | | Authors: | Luckhurst, C.A, Breccia, P, Stott, A.J, Aziz, O, Birch, H, Burli, R.W, Hughes, S, Jarvis, R.E, Lamers, M, Leonard, P, Matthews, K.L, McAllister, G, Pollack, S, Saville-Stones, E, Wishart, G, Yates, D, Dominguez, C. | | Deposit date: | 2015-05-22 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Potent, Selective, and Cns-Penetrant Tetrasubstituted Cyclopropane Class Iia Histone Deacetylase (Hdac) Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
2WBE
 
 | | Kinesin-5-Tubulin Complex with AMPPNP | | Descriptor: | BIPOLAR KINESIN KRP-130, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bodey, A.J, Kikkawa, M, Moores, C.A. | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | 9-Angstrom Structure of a Microtubule-Bound Mitotic Motor.
J.Mol.Biol., 388, 2009
|
|
2WG3
 
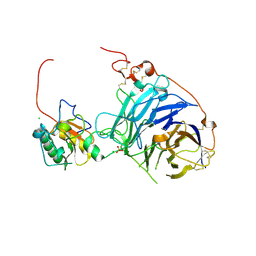 | | Crystal structure of the complex between human hedgehog-interacting protein HIP and desert hedgehog without calcium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DESERT HEDGEHOG PROTEIN N-PRODUCT, ... | | Authors: | Bishop, B, Aricescu, A.R, Harlos, K, O'Callaghan, C.A, Jones, E.Y, Siebold, C. | | Deposit date: | 2009-04-15 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into Hedgehog Ligand Sequestration by the Human Hedgehog-Interacting Protein Hip
Nat.Struct.Mol.Biol., 16, 2009
|
|
5D9H
 
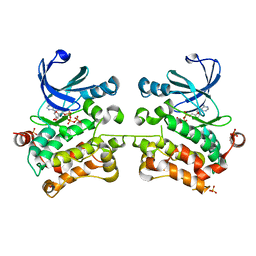 | | Crystal structure of SPAK (STK39) dimer in the basal activity state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, STE20/SPS1-related proline-alanine-rich protein kinase, ... | | Authors: | Taylor, C.A, Juang, Y.C, Goldsmith, E.J, Cobb, M.H. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Domain-Swapping Switch Point in Ste20 Protein Kinase SPAK.
Biochemistry, 54, 2015
|
|
5WNW
 
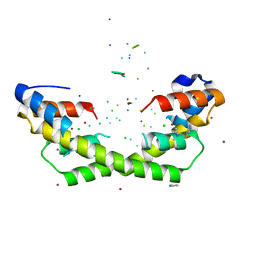 | | Chaperone Spy bound to Im7 6-45 ensemble | | Descriptor: | CHLORIDE ION, Colicin-E7 immunity protein, IMIDAZOLE, ... | | Authors: | Horowitz, S, Salmon, L, Koldewey, P, Ahlstrom, L.S, Martin, R, Xu, Q, Afonine, P.V, Trievel, R.C, Brooks, C.L, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
1TS2
 
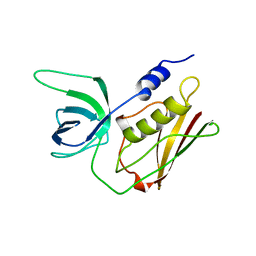 | | T128A MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-09 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
2WFT
 
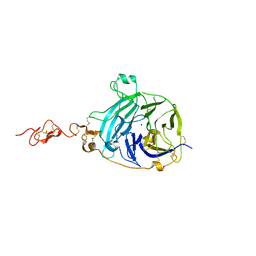 | | Crystal structure of the human HIP ectodomain | | Descriptor: | CHLORIDE ION, HEDGEHOG-INTERACTING PROTEIN, SODIUM ION | | Authors: | Bishop, B, Aricescu, A.R, Harlos, K, O'Callaghan, C.A, Jones, E.Y, Siebold, C. | | Deposit date: | 2009-04-15 | | Release date: | 2009-06-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into Hedgehog Ligand Sequestration by the Human Hedgehog-Interacting Protein Hip
Nat.Struct.Mol.Biol., 16, 2009
|
|
8FL5
 
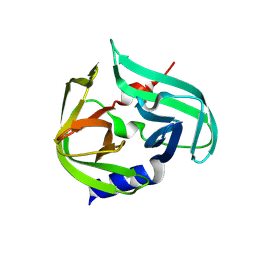 | |
4OM9
 
 | | X-Ray Crystal Structure of the passenger domain of Plasmid encoded toxin, an Autrotansporter Enterotoxin from enteroaggregative Escherichia coli (EAEC) | | Descriptor: | Serine protease pet | | Authors: | Meza-Aguilar, J.D, Fromme, P, Torres-Larios, A, Mendoza-Hernandez, G, Hernandez-Chinas, U, Arreguin-Espinosa de Los Monteros, R.A, Eslava-Campos, C.A, Fromme, R. | | Deposit date: | 2014-01-27 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of the passenger domain of plasmid encoded toxin(Pet), an autotransporter enterotoxin from enteroaggregative Escherichia coli (EAEC).
Biochem.Biophys.Res.Commun., 445, 2014
|
|
5DJE
 
 | | Crystal structure of the zuotin homology domain (ZHD) from yeast Zuo1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shrestha, O.K, Bingman, C.A, Craig, E.A. | | Deposit date: | 2015-09-02 | | Release date: | 2016-09-28 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dual interaction of the Hsp70 J-protein cochaperone Zuotin with the 40S and 60S ribosomal subunits.
Nat.Struct.Mol.Biol., 23, 2016
|
|
1RP4
 
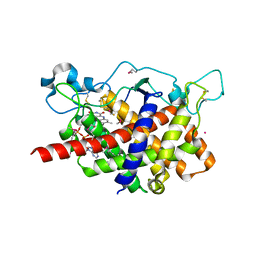 | | Structure of Ero1p, Source of Disulfide Bonds for Oxidative Protein Folding in the Cell | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, CADMIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Gross, E, Kastner, D.B, Kaiser, C.A, Fass, D. | | Deposit date: | 2003-12-03 | | Release date: | 2004-06-08 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of ero1p, source of disulfide bonds for oxidative protein folding in the cell.
Cell(Cambridge,Mass.), 117, 2004
|
|
6T2N
 
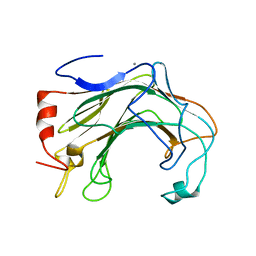 | | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown | | Descriptor: | CALCIUM ION, Glycoside hydrolase family 16 protein | | Authors: | Crouch, L.I, Liberato, M.V, Ubranowicz, P.A, Basle, A, Lamb, C.A, Cooke, K, Doona, M, Needham, S, Brady, R.R, Berrington, J.E, Madubic, K, Chater, P, Zhang, F, Linhardt, R.J, Spence, D.I.R, Bolam, D.N. | | Deposit date: | 2019-10-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown.
Nat Commun, 11, 2020
|
|
5AJ2
 
 | | Cryo electron tomography of the Naip5-Nlrc4 inflammasome | | Descriptor: | NLR FAMILY CARD DOMAIN-CONTAINING PROTEIN 4 | | Authors: | Diebolder, C.A, Halff, E.F, Koster, A.J, Huizinga, E.G, Koning, R.I. | | Deposit date: | 2015-02-20 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (40 Å) | | Cite: | Cryoelectron Tomography of the Naip5/Nlrc4 Inflammasome: Implications for Nlr Activation.
Structure, 23, 2015
|
|
