9EUU
 
 | | Structure of recombinant alpha-synuclein fibrils 1B capable of seeding GCIs in vivo | | Descriptor: | Alpha-synuclein | | Authors: | Burger, D, Kashyrina, M, Lewis, A, De Nuccio, F, Mohammed, I, de La Seigliere, H, van den Heuvel, L, Feuillie, C, Verchere, J, Berbon, M, Arotcarena, M, Retailleau, A, Bezard, E, Laferriere, F, Loquet, A, Bousset, L, Baron, T, Lofrumento, D.D, De Giorgi, F, Stahlberg, H, Ichas, F. | | Deposit date: | 2024-03-28 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Multiple System Atrophy: Insights from aSyn Fibril Structure
To Be Published
|
|
5QU1
 
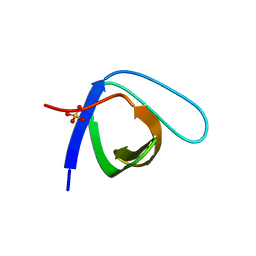 | | Crystal Structure of the monomeric human Nck SH3.1 domain, triclinic, 1.08A | | Descriptor: | Cytoplasmic protein NCK1, SULFATE ION | | Authors: | Burger, D, Ruf, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU5
 
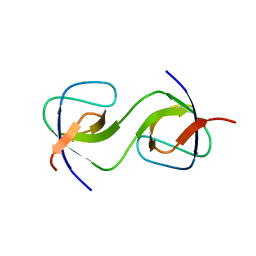 | | Domain Swap in the first SH3 domain of human Nck1 | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Burger, D, Ruf, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
3ZKX
 
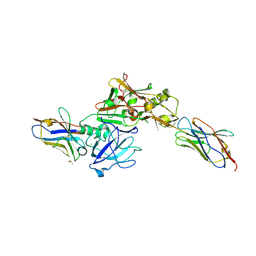 | | TERNARY BACE2 XAPERONE COMPLEX | | Descriptor: | BETA-SECRETASE 2, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kuglstatter, A, Banner, D.W, Benz, J, Bertschinger, J, Burger, D, Cuppuleri, S, Debulpaep, M, Gast, A, Grabulovski, D, Gsell, B, Hilpert, H, Huber, W, Kusznir, E, Laeremans, T, Matile, H, Rufer, A, Schlatter, D, Steyeart, J, Stihle, M, Thoma, R, Weber, M, Ruf, A. | | Deposit date: | 2013-01-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3H52
 
 | | Crystal structure of the antagonist form of human glucocorticoid receptor | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Benz, J, D'Arcy, B, Stihle, M, Burger, D, Thoma, R, Ruf, A. | | Deposit date: | 2009-04-21 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular switch in the glucocorticoid receptor: active and passive antagonist conformations
J.Mol.Biol., 395, 2010
|
|
5IO9
 
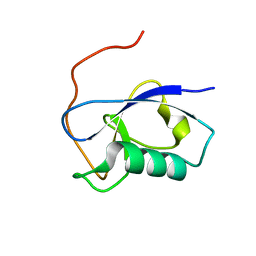 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IN7
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IP4
 
 | | X-RAY STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin, XA4551 NANOBODY AGAINST C-DCX | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IOI
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
4EYW
 
 | | Crystal structure of rat carnitine palmitoyltransferase 2 in complex with 1-[(R)-2-(3,4-Dihydro-1H-isoquinoline-2-carbonyl)-piperidin-1-yl]-2-phenoxy-ethanone | | Descriptor: | 1-[(2R)-2-(3,4-dihydroisoquinolin-2(1H)-ylcarbonyl)piperidin-1-yl]-2-phenoxyethanone, Carnitine O-palmitoyltransferase 2, mitochondrial, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Rufer, A, Joseph, C. | | Deposit date: | 2012-05-02 | | Release date: | 2013-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | Isothermal titration calorimetry with micelles: Thermodynamics of inhibitor binding to carnitine palmitoyltransferase 2 membrane protein.
FEBS Open Bio, 3, 2013
|
|
5CSZ
 
 | | CRYSTAL STRUCTURE OF GANTENERUMAB FAB FRAGMENT IN COMPLEX WITH ABETA 1-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amyloid beta A4 protein, GANTENERUMAB FAB FRAGMENT HEAVY CHAIN, ... | | Authors: | Benz, J, Burger, D, Loetscher, H.R, Bohrmann, B. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gantenerumab: a novel human anti-Abeta antibody demonstrates sustained cerebral amyloid-Beta binding and elicits cell-mediated removal of human amyloid-Beta.
J. Alzheimers Dis., 28, 2012
|
|
5Q0K
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0R
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N,1-dibenzyl-6-[(2-fluorophenyl)sulfonyl]-4,5,6,7-tetrahydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1I
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 3-(2-chlorophenyl)-N-[(1R)-1-(naphthalen-2-yl)ethyl]-5-(propan-2-yl)-1,2-oxazole-4-carboxamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0I
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 2-[(3,4-dimethoxyphenyl)-(4-methylphenyl)sulfonyl-amino]-N-(2,4-dimethylpentan-3-yl)ethanamide, Bile acid receptor, COACTIVATOR PEPTIDE PGC-1A PPAR GAMMA COACTIVATOR | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0W
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-({5-bromo-1'-[(2-chlorophenyl)sulfonyl]-2-oxospiro[indole-3,4'-piperidin]-1(2H)-yl}methyl)benzoic acid, Bile acid receptor, cDNA FLJ76652, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q12
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 2-(2,6-difluorophenyl)-N-(2,6-dimethylphenyl)-5-methylimidazo[1,2-a]pyridin-3-amine, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1G
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2E)-N-cyclohexyl-N-(cyclohexylcarbamoyl)-3-(4-fluorophenyl)prop-2-enamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1E
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 5-bromo-1-{[4-(1H-tetrazol-5-yl)phenyl]methyl}-1'-(thiophene-2-sulfonyl)spiro[indole-3,4'-piperidin]-2(1H)-one, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q10
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N-[3-(acetylamino)phenyl]-4-chloro-N-[(1S)-1-cyclohexyl-2-(cyclohexylamino)-2-oxoethyl]benzamide | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q17
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-({5-bromo-2-oxo-1'-[(thiophen-2-yl)sulfonyl]spiro[indole-3,4'-piperidin]-1(2H)-yl}methyl)benzoic acid, Bile acid receptor, Nuclear receptor coactivator peptide SRC2 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0X
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 6-(4-{[3-(3,5-dichloropyridin-4-yl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-2-methylphenyl)-1-methyl-1H-indole-3-carbox ylic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0T
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 2-phenyl-N-(propan-2-yl)-6-[(thiophen-2-yl)sulfonyl]-4,5,6,7-tetrahydro-1H-pyrrolo[2,3-c]pyridine-1-carboxamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q11
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N,N-dicyclohexyl-3-(2,4-dichlorophenyl)-5-methyl-1,2-oxazole-4-carboxamide | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0V
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexyl-N-(2-fluorophenyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
