3ZOP
 
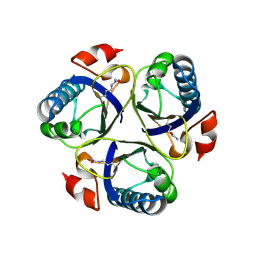 | | Arg90Cit chorismate mutase of Bacillus subtilis at 1.6 A resolution | | Descriptor: | CHORISMATE MUTASE AROH | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-22 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZP4
 
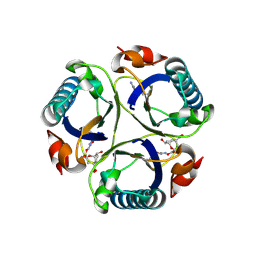 | | Arg90Cit chorismate mutase of Bacillus subtilis in complex with a transition state analog | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, CHORISMATE MUTASE AROH | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-26 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZP7
 
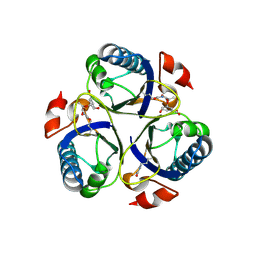 | | Arg90Cit chorismate mutase of Bacillus subtilis in complex with chorismate and prephenate | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, CHORISMATE MUTASE AROH, PREPHENIC ACID | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-26 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1HRM
 
 | |
8ASJ
 
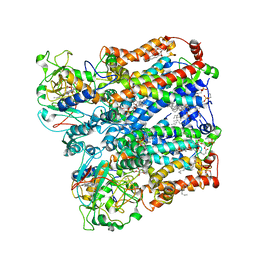 | | Four subunit cytochrome b-c1 complex from Rhodobacter sphaeroides in native nanodiscs - focussed refinement in the b-c conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Cytochrome b, Cytochrome b-c1 subunit IV, ... | | Authors: | Swainsbury, D.J.K, Hawkings, F.R, Martin, E.C, Musial, S, Salisbury, J.H, Jackson, P.J, Farmer, D.A, Johnson, M.P, Siebert, C.A, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2022-08-19 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structure of the four-subunit Rhodobacter sphaeroides cytochrome bc 1 complex in styrene maleic acid nanodiscs.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ASI
 
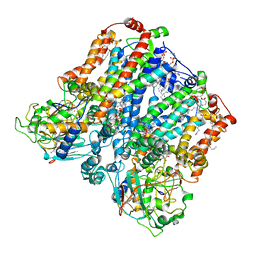 | | Four subunit cytochrome b-c1 complex from Rhodobacter sphaeroides in native nanodiscs - consensus refinement in the b-b conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Cytochrome b, Cytochrome b-c1 subunit IV, ... | | Authors: | Swainsbury, D.J.K, Hawkings, F.R, Martin, E.C, Musial, S, Salisbury, J.H, Jackson, P.J, Farmer, D.A, Johnson, M.P, Siebert, C.A, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2022-08-19 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the four-subunit Rhodobacter sphaeroides cytochrome bc 1 complex in styrene maleic acid nanodiscs.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5UTG
 
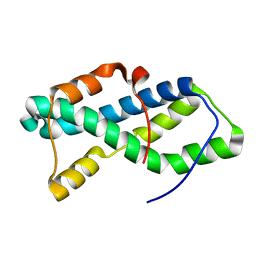 | | Red abalone lysin F104A | | Descriptor: | Egg-lysin | | Authors: | Wilburn, D.B, Tuttle, L.M. | | Deposit date: | 2017-02-14 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of sperm lysin yields novel insights into molecular dynamics of rapid protein evolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5QU1
 
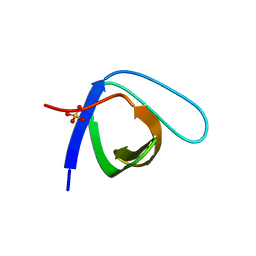 | | Crystal Structure of the monomeric human Nck SH3.1 domain, triclinic, 1.08A | | Descriptor: | Cytoplasmic protein NCK1, SULFATE ION | | Authors: | Burger, D, Ruf, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU5
 
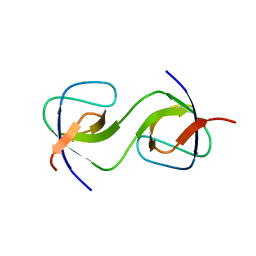 | | Domain Swap in the first SH3 domain of human Nck1 | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Burger, D, Ruf, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
3FC6
 
 | | hRXRalpha & mLXRalpha with an indole Pharmacophore, SB786875 | | Descriptor: | Nr1h3 protein, RETINOIC ACID, Retinoic acid receptor RXR-alpha, ... | | Authors: | Washburn, D.G, Hoang, T.H, Campobasso, N, Smallwood, A, Parks, D.J, Webb, C.L, Frank, K, Nord, M, Duraiswami, C, Evans, C, Jaye, M, Thompson, S.K. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | Synthesis and SAR of potent LXR agonists containing an indole pharmacophore.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1AWO
 
 | |
1J7I
 
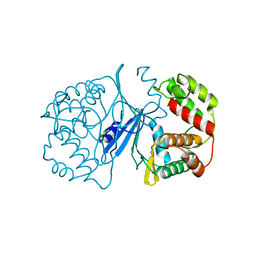 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa Apoenzyme | | Descriptor: | AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
2MHY
 
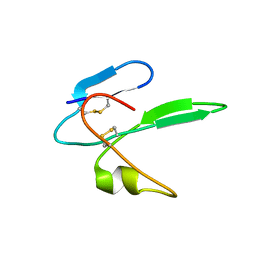 | | Structure determination of the salamander courtship pheromone Plethodontid Modulating Factor | | Descriptor: | Plethodontid modulating factor | | Authors: | Wilburn, D.B, Bowen, K.E, Doty, K.A, Arumugam, S, Lane, A.N, Feldhoff, P.W, Feldhoff, R.C. | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the evolution of a sexy protein: novel topology and restricted backbone flexibility in a hypervariable pheromone from the red-legged salamander, Plethodon shermani.
Plos One, 9, 2014
|
|
5HUB
 
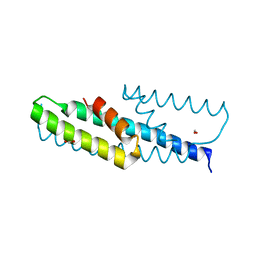 | | High-resolution structure of chorismate mutase from Corynebacterium glutamicum | | Descriptor: | Chorismate mutase, FORMIC ACID | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
2KWU
 
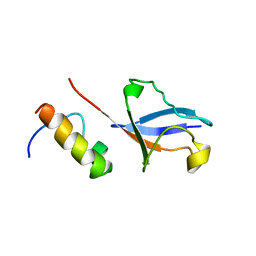 | | Solution Structure of UBM2 of murine Polymerase iota in Complex with Ubiquitin | | Descriptor: | DNA polymerase iota, Ubiquitin | | Authors: | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | Deposit date: | 2010-04-19 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2KWV
 
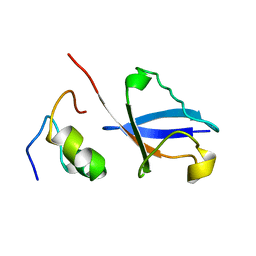 | | Solution Structure of UBM1 of murine Polymerase iota in Complex with Ubiquitin | | Descriptor: | DNA polymerase iota, Ubiquitin | | Authors: | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | Deposit date: | 2010-04-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
1J7U
 
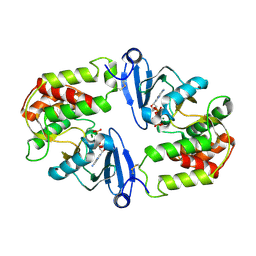 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa AMPPNP Complex | | Descriptor: | AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
1J7L
 
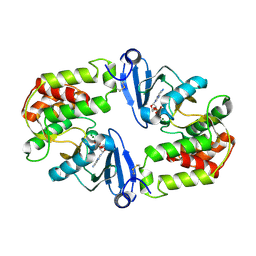 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa ADP Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE, MAGNESIUM ION | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-17 | | Release date: | 2001-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
4TR6
 
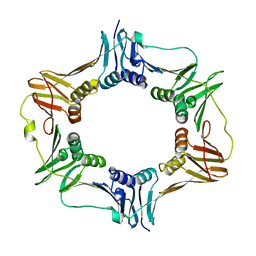 | | Crystal structure of DNA polymerase sliding clamp from Bacillus subtilis | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION | | Authors: | Burnouf, D, Olieric, V, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-14 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
5HUC
 
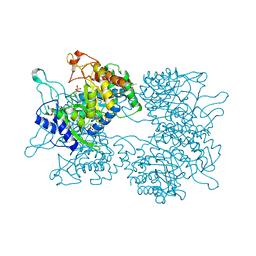 | | DAHP synthase from Corynebacterium glutamicum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-Deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, MANGANESE (II) ION, ... | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
5HUE
 
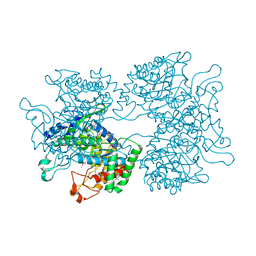 | | DAHP synthase from Corynebacterium glutamicum in complex with tryptophan | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-Deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, ... | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
3ZO8
 
 | | Wild-type chorismate mutase of Bacillus subtilis at 1.6 A resolution | | Descriptor: | CHORISMATE MUTASE AROH | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-20 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6AZ5
 
 | |
4UF2
 
 | | Deerpox virus DPV022 in complex with Bax BH3 | | Descriptor: | Antiapoptotic membrane protein, Apoptosis regulator BAX | | Authors: | Burton, D.R, Kvansakul, M. | | Deposit date: | 2014-12-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Deerpox Virus-Mediated Inhibition of Apoptosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UF3
 
 | | Deerpox virus DPV022 in complex with Bim BH3 | | Descriptor: | Antiapoptotic membrane protein, Bcl-2-like protein 11 | | Authors: | Burton, D.R, Kvansakul, M. | | Deposit date: | 2014-12-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Deerpox Virus-Mediated Inhibition of Apoptosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
