5Q0S
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-N-(2-cyanophenyl)-2-cyclohexylacetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0J
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-N,2-dicyclohexyl-2-[2-(5-phenylthiophen-2-yl)-1H-benzimidazol-1-yl]acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
2Q99
 
 | |
6U6N
 
 | | Structure of the trimeric globular domain of Adiponectin mutant - D187A Q188A | | 分子名称: | Adiponectin, CHLORIDE ION | | 著者 | Pascolutti, R, Kruse, A.C, Erlandson, S.C, Burri, D.J, Zheng, S. | | 登録日 | 2019-08-30 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Mapping and engineering the interaction between adiponectin and T-cadherin.
J.Biol.Chem., 295, 2020
|
|
3KBA
 
 | | Progesterone receptor bound to sulfonamide pyrrolidine partial agonist | | 分子名称: | 2-chloro-4-{(2-methylbenzyl)[(3S)-1-(methylsulfonyl)pyrrolidin-3-yl]amino}benzonitrile, Progesterone receptor, SULFATE ION | | 著者 | Kallander, L.S, Washburn, D.G, Williams, S.P, Madauss, K.P. | | 登録日 | 2009-10-20 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Improving the developability profile of pyrrolidine progesterone receptor partial agonists.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6U66
 
 | | Structure of the trimeric globular domain of Adiponectin | | 分子名称: | Adiponectin, CALCIUM ION, SODIUM ION | | 著者 | Pascolutti, R, Kruse, A.C, Erlandson, S.C, Burri, D.J, Zheng, S. | | 登録日 | 2019-08-29 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (0.99 Å) | | 主引用文献 | Mapping and engineering the interaction between adiponectin and T-cadherin.
J.Biol.Chem., 295, 2020
|
|
1QI9
 
 | | X-RAY SIRAS STRUCTURE DETERMINATION OF A VANADIUM-DEPENDENT HALOPEROXIDASE FROM ASCOPHYLLUM NODOSUM AT 2.0 A RESOLUTION | | 分子名称: | VANADATE ION, Vanadium-dependent bromoperoxidase | | 著者 | Weyand, M, Hecht, H.-J, Kiess, M, Liaud, M.F, Vilter, H, Schomburg, D. | | 登録日 | 1999-06-10 | | 公開日 | 2000-06-10 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | X-ray structure determination of a vanadium-dependent haloperoxidase from Ascophyllum nodosum at 2.0 A resolution.
J.Mol.Biol., 293, 1999
|
|
3ZKX
 
 | | TERNARY BACE2 XAPERONE COMPLEX | | 分子名称: | BETA-SECRETASE 2, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Kuglstatter, A, Banner, D.W, Benz, J, Bertschinger, J, Burger, D, Cuppuleri, S, Debulpaep, M, Gast, A, Grabulovski, D, Gsell, B, Hilpert, H, Huber, W, Kusznir, E, Laeremans, T, Matile, H, Rufer, A, Schlatter, D, Steyeart, J, Stihle, M, Thoma, R, Weber, M, Ruf, A. | | 登録日 | 2013-01-25 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1QW9
 
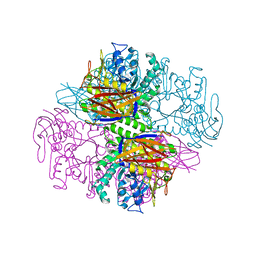 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with 4-nitrophenyl-Ara | | 分子名称: | 4-nitrophenyl alpha-L-arabinofuranoside, Alpha-L-arabinofuranosidase | | 著者 | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | 登録日 | 2003-09-01 | | 公開日 | 2003-10-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1QQ9
 
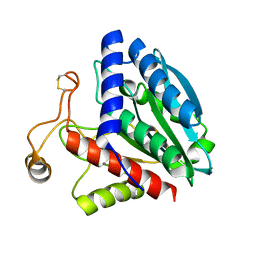 | | STREPTOMYCES GRISEUS AMINOPEPTIDASE COMPLEXED WITH METHIONINE | | 分子名称: | AMINOPEPTIDASE, CALCIUM ION, METHIONINE, ... | | 著者 | Gilboa, R, Greenblatt, H.M, Perach, M, Spungin-Bialik, A, Lessel, U, Schomburg, D, Blumberg, S, Shoham, G. | | 登録日 | 1999-06-12 | | 公開日 | 2000-05-03 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Interactions of Streptomyces griseus aminopeptidase with a methionine product analogue: a structural study at 1.53 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QW8
 
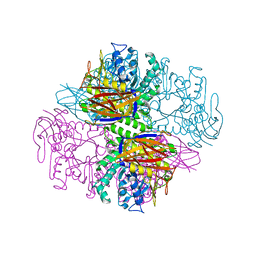 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with Ara-alpha(1,3)-Xyl | | 分子名称: | Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose | | 著者 | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | 登録日 | 2003-09-01 | | 公開日 | 2003-10-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
7VKV
 
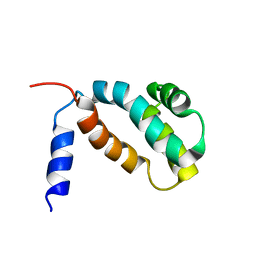 | |
8FG1
 
 | | Human diaphanous inhibitory domain bound to diaphanous autoregulatory domain | | 分子名称: | Protein diaphanous homolog 1 | | 著者 | Ramirez, L.M.S, Theophall, G, Premo, A, Manigrasso, M, Yepuri, G, Burz, D, Ramasamy, R, Schmidt, A.M, Shekhtman, A. | | 登録日 | 2022-12-12 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Disruption of the productive encounter complex results in dysregulation of DIAPH1 activity.
J.Biol.Chem., 299, 2023
|
|
1JEG
 
 | | Solution structure of the SH3 domain from C-terminal Src Kinase complexed with a peptide from the tyrosine phosphatase PEP | | 分子名称: | HEMATOPOIETIC CELL PROTEIN-TYROSINE PHOSPHATASE 70Z-PEP, TYROSINE-PROTEIN KINASE CSK | | 著者 | Ghose, R, Shekhtman, A, Goger, M.J, Ji, H, Cowburn, D. | | 登録日 | 2001-06-17 | | 公開日 | 2001-10-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel, specific interaction involving the Csk SH3 domain and its natural ligand.
Nat.Struct.Biol., 8, 2001
|
|
3JYP
 
 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with quinate and NADH | | 分子名称: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | 著者 | Hoeppner, A, Schomburg, D, Niefind, K. | | 登録日 | 2009-09-22 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
3JYO
 
 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with NAD | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | 著者 | Hoeppner, A, Niefind, K, Schomburg, D. | | 登録日 | 2009-09-22 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
3JYQ
 
 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with shikimate and NADH | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | 著者 | Hoeppner, A, Schomburg, D, Niefind, K. | | 登録日 | 2009-09-22 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
5JBT
 
 | | Mesotrypsin in complex with cleaved amyloid precursor like protein 2 inhibitor (APLP2) | | 分子名称: | Amyloid-like protein 2, CALCIUM ION, PRSS3 protein, ... | | 著者 | Kayode, O, Wang, R, Pendlebury, D, Soares, A, Radisky, E.S. | | 登録日 | 2016-04-13 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | An Acrobatic Substrate Metamorphosis Reveals a Requirement for Substrate Conformational Dynamics in Trypsin Proteolysis.
J. Biol. Chem., 291, 2016
|
|
5TVZ
 
 | | Solution NMR structure of Saccharomyces cerevisiae Pom152 Ig-like repeat, residues 718-820 | | 分子名称: | Nucleoporin POM152 | | 著者 | Dutta, K, Sampathkumar, P, Cowburn, D, Almo, S.C, Rout, M.P, Fernandez-Martinez, J. | | 登録日 | 2016-11-10 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure, 25, 2017
|
|
1AWE
 
 | |
1G84
 
 | | THE SOLUTION STRUCTURE OF THE C EPSILON2 DOMAIN FROM IGE | | 分子名称: | IMMUNOGLOBULIN E | | 著者 | McDonnell, J.M, Cowburn, D, Gould, H.J, Sutton, B.J, Calvert, R, Beavil, R.E, Beavil, A.J, Henry, A.J. | | 登録日 | 2000-11-16 | | 公開日 | 2001-05-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of the IgE Cepsilon2 domain and its role in stabilizing the complex with its high-affinity receptor FcepsilonRIalpha.
Nat.Struct.Biol., 8, 2001
|
|
1CX1
 
 | | SECOND N-TERMINAL CELLULOSE-BINDING DOMAIN FROM CELLULOMONAS FIMI BETA-1,4-GLUCANASE C, NMR, 22 STRUCTURES | | 分子名称: | ENDOGLUCANASE C | | 著者 | Brun, E, Johnson, P.E, Creagh, L.A, Haynes, C.A, Tomme, P, Webster, P, Kilburn, D.G, McIntosh, L.P. | | 登録日 | 1999-08-27 | | 公開日 | 2000-04-02 | | 最終更新日 | 2024-10-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and binding specificity of the second N-terminal cellulose-binding domain from Cellulomonas fimi endoglucanase C.
Biochemistry, 39, 2000
|
|
3BRH
 
 | |
6AX5
 
 | | RPT1 region of INI1/SNF5/SMARCB1_HUMAN - SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1. | | 分子名称: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | 著者 | Girvin, M.E, Cahill, S.M, Harris, R, Cowburn, D, Spira, M, Wu, X, Prakash, R, Bernowitz, M, Almo, S.C, Kalpana, G.V. | | 登録日 | 2017-09-06 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | INI1/SMARCB1 Rpt1 domain mimics TAR RNA in binding to integrase to facilitate HIV-1 replication.
Nat Commun, 12, 2021
|
|
3EAZ
 
 | |
