2X5I
 
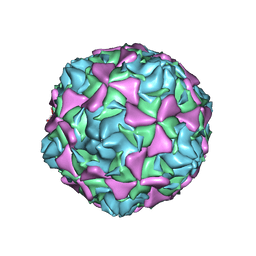 | | Crystal structure echovirus 7 | | Descriptor: | LAURIC ACID, VP1, VP2, ... | | Authors: | Plevka, P, Hafenstein, S, Zhang, Y, Bowman, V.D, Chipman, P.R, Bator, C.M, Rossmann, M.G. | | Deposit date: | 2010-02-08 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Interaction of Decay-Accelerating Factor with Echovirus 7.
J.Virol., 84, 2010
|
|
1M06
 
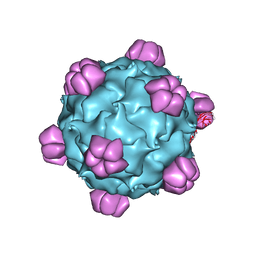 | | Structural Studies of Bacteriophage alpha3 Assembly, X-Ray Crystallography | | Descriptor: | 5'-D(P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR))-3', Capsid Protein, Major spike protein, ... | | Authors: | Bernal, R.A, Hafenstein, S, Olson, N.H, Bowman, V, Chipman, P.R, Baker, T.S, Fane, B.A, Rossmann, M.G. | | Deposit date: | 2002-06-12 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Studies of Bacteriophage alpha3 Assembly
J.Mol.Biol., 325, 2003
|
|
3GK8
 
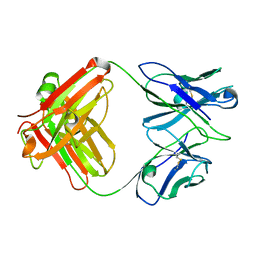 | | X-ray crystal structure of the Fab from MAb 14, mouse antibody against Canine Parvovirus | | Descriptor: | Fab 14 Heavy Chain, Fab 14 Light Chain | | Authors: | Hafenstein, S, Bowman, V, Sun, T, Nelson, C, Palermo, L, Chipman, P, Battisti, A, Parrish, C. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids.
J.Virol., 83, 2009
|
|
3URO
 
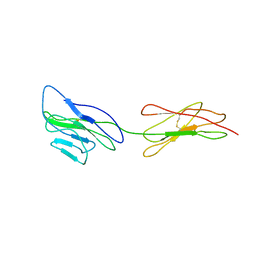 | | Poliovirus receptor CD155 D1D2 | | Descriptor: | Poliovirus receptor | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-07 | | Method: | X-RAY DIFFRACTION (3.5005 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3J2M
 
 | | The X-ray structure of the gp15 hexamer and the model of the gp18 protein fitted into the cryo-EM reconstruction of the extended T4 tail | | Descriptor: | Tail connector protein Gp15, Tail sheath protein Gp18 | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-09 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
3J2N
 
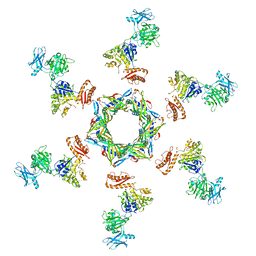 | | The X-ray structure of the gp15 hexamer and the model of the gp18 protein fitted into the cryo-EM reconstruction of the contracted T4 tail | | Descriptor: | Tail connector protein Gp15, Tail sheath protein Gp18 | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-10 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
4HUH
 
 | | Structure of the bacteriophage T4 tail terminator protein, gp15 (C-terminal truncation mutant 1-261). | | Descriptor: | Tail connector protein Gp15 | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-02 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
3J2O
 
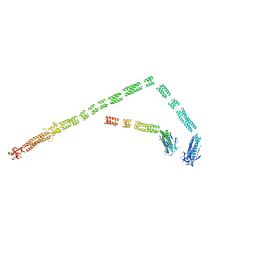 | | Model of the bacteriophage T4 fibritin based on the cryo-EM reconstruction of the contracted T4 tail containing the phage collar and whiskers | | Descriptor: | Fibritin | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A.A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-12 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
4HUD
 
 | | Structure of the bacteriophage T4 tail terminator protein, gp15. | | Descriptor: | Tail connector protein Gp15 | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7001 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
3SHS
 
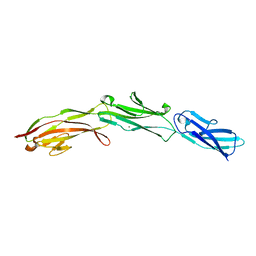 | | Three N-terminal domains of the bacteriophage RB49 Highly Immunogenic Outer Capsid protein (Hoc) | | Descriptor: | Hoc head outer capsid protein, MAGNESIUM ION | | Authors: | Fokine, A, Islam, M.Z, Zhang, Z, Bowman, V.D, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2011-06-16 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage.
J.Virol., 85, 2011
|
|
3IY0
 
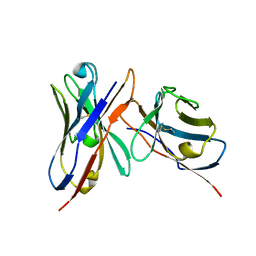 | | Variable domains of the x-ray structure of Fab 14 fitted into the cryoEM reconstruction of the virus-Fab 14 complex | | Descriptor: | Fab 14, heavy domain, light domain | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-07 | | Release date: | 2009-05-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
1M0F
 
 | | Structural Studies of Bacteriophage alpha3 Assembly, Cryo-electron microscopy | | Descriptor: | Capsid Protein F, Major Spike Protein G, Scaffolding Protein B, ... | | Authors: | Bernal, R.A, Hafenstein, S, Olson, N.H, Bowman, V.D, Chipman, P.R, Baker, T.S, Fane, B.A, Rossmann, M.G. | | Deposit date: | 2002-06-12 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structural Studies of Bacteriophage alpha3 Assembly
J.Mol.Biol., 325, 2003
|
|
3IY7
 
 | | Variable domains of the computer generated model (WAM) of Fab F fitted into the cryoEM reconstruction of the virus-Fab F complex | | Descriptor: | fragment from neutralizing antibody F (heavy chain), fragment from neutralizing antibody F (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY2
 
 | | Variable domains of the computer generated model (WAM) of Fab 6 fitted into the cryoEM reconstruction of the virus-Fab 6 complex | | Descriptor: | Antibody 6, heavy chain, light chain | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY4
 
 | | Variable domains of the computer generated model (WAM) of Fab 15 fitted into the cryoEM reconstruction of the virus-Fab 15 complex | | Descriptor: | fragment of neutralizing antibody 15 (heavy chain), fragment of neutralizing antibody 15 (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IYP
 
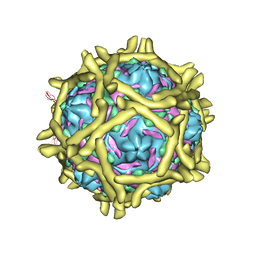 | | The Interaction of Decay-accelerating Factor with Echovirus 7 | | Descriptor: | Capsid protein, Complement decay-accelerating factor, LAURIC ACID, ... | | Authors: | Plevka, P, Hafenstein, S, Zhang, Y, Harris, K.G, Cifuente, J.O, Bowman, V.D, Chipman, P.R, Lin, F, Medof, D.E, Bator, C.M, Rossmann, M.G. | | Deposit date: | 2010-04-07 | | Release date: | 2010-11-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Interaction of decay-accelerating factor with echovirus 7.
J.Virol., 84, 2010
|
|
3IY6
 
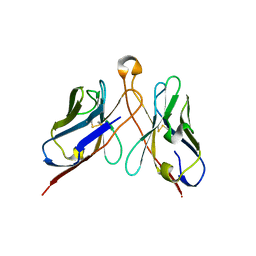 | | Variable domains of the computer generated model (WAM) of Fab E fitted into the cryoEM reconstruction of the virus-Fab E complex | | Descriptor: | fragment from neutralizing antibody E (heavy chain), fragment from neutralizing antibody E (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY1
 
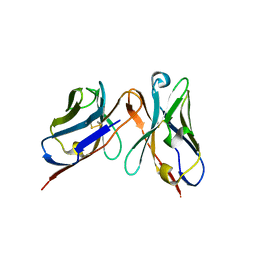 | | Variable domains of the WAM of Fab B fitted into the cryoEM reconstruction of the virus-Fab B complex | | Descriptor: | Fab B, heavy chain, light chain | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY5
 
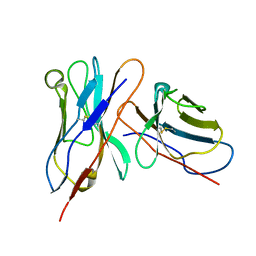 | | Variable domains of the mouse Fab (1AIF) fitted into the cryoEM reconstruction of the virus-Fab 16 complex | | Descriptor: | antibody fragment IGG2A (heavy chain), antibody fragment IGG2A (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY3
 
 | | Variable domains of the computer generated model (WAM) of Fab 8 fitted into the cryoEM reconstruction of the virus-Fab 8 complex | | Descriptor: | antibody fragment from neutralizing antibody 8 (heavy chain), antibody fragment from neutralizing antibody 8 (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (11.1 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
1NN8
 
 | | CryoEM structure of poliovirus receptor bound to poliovirus | | Descriptor: | MYRISTIC ACID, coat protein VP1, coat protein VP2, ... | | Authors: | He, Y, Mueller, S, Chipman, P.R, Bator, C.M, Peng, X, Bowman, V.D, Mukhopadhyay, S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2003-01-13 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Complexes of poliovirus serotypes with their common cellular receptor, CD155
J.Virol., 77, 2003
|
|
2QZH
 
 | | SCR2/3 of DAF from the NMR structure 1nwv fitted into a cryoEM reconstruction of CVB3-RD complexed with DAF | | Descriptor: | Complement decay-accelerating factor | | Authors: | Hafenstein, S, Bowman, V.D, Chipman, P.R, Bator Kelly, C.M, Lin, F, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2007-08-16 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Interaction of decay-accelerating factor with coxsackievirus b3.
J.Virol., 81, 2007
|
|
2QZD
 
 | | Fitted structure of SCR4 of DAF into cryoEM density | | Descriptor: | Complement decay-accelerating factor | | Authors: | Hafenstein, S, Bowman, V.D, Chipman, P.R, Bator Kelly, C.M, Lin, F, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2007-08-16 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Interaction of decay-accelerating factor with coxsackievirus b3.
J.Virol., 81, 2007
|
|
2QZF
 
 | | SCR1 of DAF from 1ojv fitted into cryoEM density | | Descriptor: | Complement decay-accelerating factor | | Authors: | Hafenstein, S, Bowman, V.D, Chipman, P.R, Bator Kelly, C.M, Lin, F, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2007-08-16 | | Release date: | 2007-10-30 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Interaction of decay-accelerating factor with coxsackievirus b3.
J.Virol., 81, 2007
|
|
3EPD
 
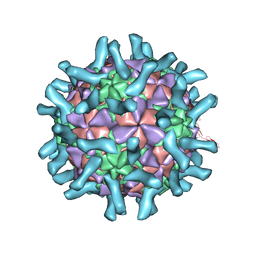 | | CryoEM structure of poliovirus receptor bound to poliovirus type 3 | | Descriptor: | MYRISTIC ACID, Poliovirus Type3 peptide, Poliovirus receptor, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
