5Q48
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 73) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4IH8
 
 | | Crystal structure of TgCDPK1 with inhibitor bound | | Descriptor: | 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane, Calmodulin-domain protein kinase 1 | | Authors: | El Bakkouri, M, Tempel, W, Crandall, I, Massad, T, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kain, K, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.877 Å) | | Cite: | Crystal structure of TgCDPK1 with inhibitor bound
To be Published
|
|
4IFG
 
 | | Crystal structure of TgCDPK1 with inhibitor bound | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, Calmodulin-domain protein kinase 1, UNKNOWN ATOM OR ION | | Authors: | El Bakkouri, M, Tempel, W, Crandall, I, Massad, T, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kain, K, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-14 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of TgCDPK1 with inhibitor bound
To be Published
|
|
4IJ8
 
 | | Crystal structure of the complex of SETD8 with SAM | | Descriptor: | N-lysine methyltransferase SETD8, S-ADENOSYLMETHIONINE, UNKNOWN ATOM OR ION, ... | | Authors: | Yu, W, Tempel, W, Li, Y, El Bakkouri, M, Shapira, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-21 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of SETD8 with SAM
To be Published
|
|
4TLG
 
 | | Crystal structure of SEC14-like protein 4 (SEC14L4) | | Descriptor: | 1,2-ETHANEDIOL, SEC14-like protein 4, UNDECANOIC ACID | | Authors: | Vollmar, M, Kopec, J, Kiyani, W, Shrestha, L, Sorrell, F, Krojer, T, Williams, E, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-05-29 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of SEC14-like protein 4 (SEC14L4)
To Be Published
|
|
4OCT
 
 | | Crystal structure of human ALKBH5 crystallized in the presence of Mn^{2+} and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, MANGANESE (II) ION, RNA demethylase ALKBH5, ... | | Authors: | Tempel, W, Chao, X, Liu, K, Dong, A, Cerovina, T, He, H, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-01-09 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation.
J.Biol.Chem., 289, 2014
|
|
5Q4Z
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 100) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q3A
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 38) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q3R
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 55) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4AVX
 
 | | Hepatocyte Growth Factor-Regulated Tyrosine Kinase Substrate (Hgs-Hrs) bound to an IP2 compound at 1.68 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, HEPATOCYTE GROWTH FACTOR-REGULATED TYROSINE KINASE SUBSTRATE, PHOSPHORIC ACID MONO-(2,3,4,6-TETRAHYDROXY-5-PHOSPHONOOXY-CYCLOHEXYL) ESTER, ... | | Authors: | Williams, E, Canning, P, Shrestha, L, Krojer, T, Vollmar, M, Slowey, A, Conway, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bullock, A. | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of the Tandem Vhs and Fyve Domains of Hepatocyte Growth Factor-Regulated Tyrosine Kinase Substrate (Hgs-Hrs) Bound to an Ip2 Compound at 1.68 A Resolution
To be Published
|
|
4DIP
 
 | | Crystal structure of human Peptidyl-prolyl cis-trans isomerase FKBP14 | | Descriptor: | PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase FKBP14, SODIUM ION | | Authors: | Krojer, T, Kiyani, W, Goubin, S, Muniz, J.R.C, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Oppermann, U, Zschocke, J, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-01-31 | | Release date: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of human Peptidyl-prolyl cis-trans isomerase FKBP14
To be Published
|
|
4ATM
 
 | | Crystal structure of the BAR domain of human Amphiphysin, isoform 1 at 1.8 Angstrom resolution featuring increased order at the N- terminus. | | Descriptor: | 1,2-ETHANEDIOL, AMPHIPHYSIN, GLYCEROL | | Authors: | Allerston, C.K, Krojer, T, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Crystal Structure of the Bar Domain of Human Amphiphysin, Isoform 1
To be Published
|
|
4BKJ
 
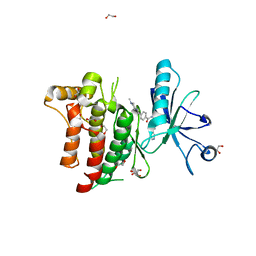 | | Crystal structure of the human DDR1 kinase domain in complex with imatinib | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Krojer, T, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Mechanisms Determining Inhibition of the Collagen Receptor Ddr1 by Selective and Multi-Targeted Type II Kinase Inhibitors
J.Mol.Biol., 426, 2014
|
|
5Q57
 
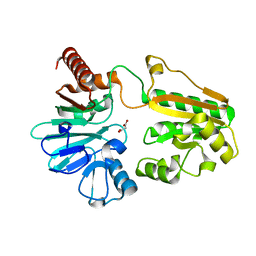 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 108) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q4I
 
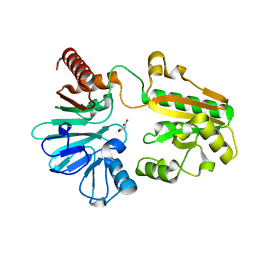 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 83) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q5J
 
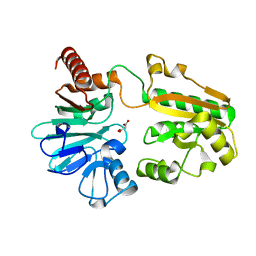 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 120) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q5Z
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 136) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q1N
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000295a | | Descriptor: | DNA cross-link repair 1A protein, MALONATE ION, NICKEL (II) ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R61
 
 | | PanDDA analysis group deposition -- Crystal Structure of FIBRINOGEN-LIKE GLOBE DOMAIN OF HUMAN TENASCIN-C in complex with Z1578665941 | | Descriptor: | 1-(3-methyl-1,2,4-thiadiazol-5-yl)-1,4-diazepane, Tenascin C (Hexabrachion), isoform CRA_a | | Authors: | Coker, J.A, Bezerra, G.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Yue, W.W, Marsden, B.D. | | Deposit date: | 2020-02-28 | | Release date: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RJI
 
 | | PanDDA analysis group deposition of ground-state model of PHIP | | Descriptor: | PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RJX
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z285782452 | | Descriptor: | N-methyl-2-(methylsulfonyl)aniline, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
3CBQ
 
 | | Crystal structure of the human REM2 GTPase with bound GDP | | Descriptor: | 1,2-ETHANEDIOL, GTP-binding protein REM 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nedyalkova, L, Shen, Y, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-22 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of the human REM2 GTPase with bound GDP.
To be Published
|
|
5RAO
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001084a | | Descriptor: | CHLORIDE ION, Lysine-specific demethylase 3B, MANGANESE (II) ION, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RB5
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM010010a | | Descriptor: | CHLORIDE ION, Lysine-specific demethylase 3B, MANGANESE (II) ION, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RKD
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z2168282707 | | Descriptor: | (6S)-1-methyl-4,5,6,7-tetrahydro-1H-benzotriazole-6-carboxylic acid, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.237 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
