5AIP
 
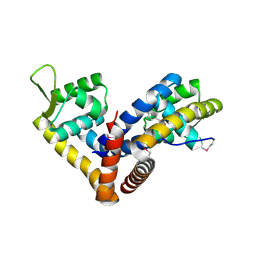 | | Crystal structure of NadR in complex with 4-hydroxyphenylacetate | | Descriptor: | 4-HYDROXYPHENYLACETATE, TRANSCRIPTIONAL REGULATOR, MARR FAMILY | | Authors: | Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2015-02-16 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis of Ligand-Dependent Regulation of Nadr, the Transcriptional Repressor of Meningococcal Virulence Factor Nada.
Plos Pathog., 12, 2016
|
|
4D7W
 
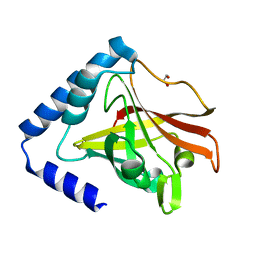 | | Crystal structure of sortase C1 (SrtC1) from Streptococcus agalactiae | | Descriptor: | 1,2-ETHANEDIOL, SORTASE FAMILY PROTEIN | | Authors: | Malito, E, Lazzarin, M, Cozzi, R, Bottomley, M.J. | | Deposit date: | 2014-11-28 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Noncanonical Sortase-Mediated Assembly of Pilus Type 2B in Group B Streptococcus.
Faseb J., 29, 2015
|
|
4UZG
 
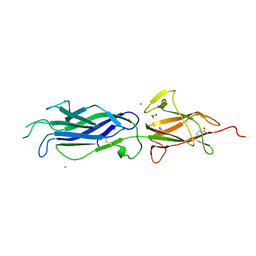 | | Crystal structure of group B streptococcus pilus 2b backbone protein SAK_1440 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SURFACE PROTEIN SPB1 | | Authors: | Malito, E, Cozzi, R, Bottomley, M.J. | | Deposit date: | 2014-09-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structure and assembly of group B streptococcus pilus 2b backbone protein.
PLoS ONE, 10, 2015
|
|
1K1G
 
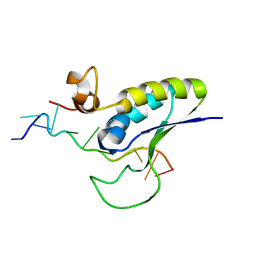 | | STRUCTURAL BASIS FOR RECOGNITION OF THE INTRON BRANCH SITE RNA BY SPLICING FACTOR 1 | | Descriptor: | 5'-R(*UP*AP*UP*AP*CP*UP*AP*AP*CP*AP*A)-3', SF1-Bo isoform | | Authors: | Liu, Z, Luyten, I, Bottomley, M.J, Messias, A.C, Houngninou-Molango, S, Sprangers, R, Zanier, K, Kramer, A, Sattler, M. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of the intron branch site RNA by splicing factor 1.
Science, 294, 2001
|
|
3P5C
 
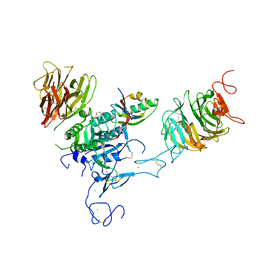 | | The structure of the LDLR/PCSK9 complex reveals the receptor in an extended conformation | | Descriptor: | CALCIUM ION, Low density lipoprotein receptor variant, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lo Surdo, P, Bottomley, M.J, Calzetta, A, Settembre, E.C, Cirillo, A, Pandit, S, Ni, Y, Hubbard, B, Sitlani, A, Carfi, A. | | Deposit date: | 2010-10-08 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH.
Embo Rep., 12, 2011
|
|
3P5B
 
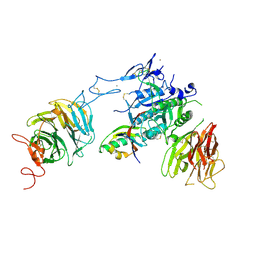 | | The structure of the LDLR/PCSK9 complex reveals the receptor in an extended conformation | | Descriptor: | CALCIUM ION, Low density lipoprotein receptor variant, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lo Surdo, P, Bottomley, M.J, Calzetta, A, Settembre, E.C, Cirillo, A, Pandit, S, Ni, Y, Hubbard, B, Sitlani, A, Carfi, A. | | Deposit date: | 2010-10-08 | | Release date: | 2011-10-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH.
Embo Rep., 12, 2011
|
|
4BIG
 
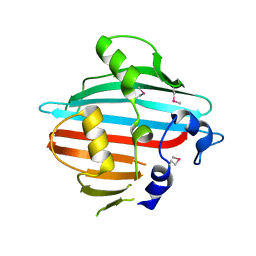 | | Crystal structure of the conserved staphylococcal antigen 1B, Csa1B | | Descriptor: | UNCHARACTERIZED LIPOPROTEIN SAOUHSC_00053 | | Authors: | Malito, E, Bottomley, M.J, Schluepen, C, Liberatori, S. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.274 Å) | | Cite: | Mining the Bacterial Unknown Proteome: Identification and Characterization of a Novel Family of Highly Conserved Protective Antigens in Staphylococcus Aureus
Biochem.J., 455, 2013
|
|
4B8Y
 
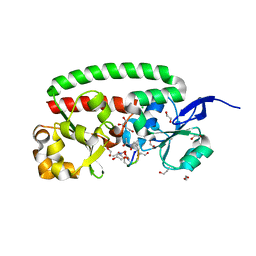 | | Ferrichrome-bound FhuD2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Malito, E, Bottomley, M.J, Spraggon, G. | | Deposit date: | 2012-08-31 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Staphylococcus Aureus Virulence Factor and Vaccine Candidate Fhud2.
Biochem.J., 449, 2013
|
|
4BIH
 
 | | Crystal structure of the conserved staphylococcal antigen 1A, Csa1A | | Descriptor: | CALCIUM ION, UNCHARACTERIZED LIPOPROTEIN SAOUHSC_00053 | | Authors: | Malito, E, Bottomley, M.J, Spraggon, G, Schluepen, C, Liberatori, S. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.459 Å) | | Cite: | Mining the Bacterial Unknown Proteome: Identification and Characterization of a Novel Family of Highly Conserved Protective Antigens in Staphylococcus Aureus
Biochem.J., 455, 2013
|
|
4CJD
 
 | | Crystal structure of Neisseria meningitidis trimeric autotransporter and vaccine antigen NadA | | Descriptor: | IODIDE ION, NADA | | Authors: | Malito, E, Biancucci, M, Spraggon, G, Bottomley, M.J. | | Deposit date: | 2013-12-19 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | Structure of the Meningococcal Vaccine Antigen Nada and Epitope Mapping of a Bactericidal Antibody.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1OQJ
 
 | | Crystal structure of the SAND domain from glucocorticoid modulatory element binding protein-1 (GMEB1) | | Descriptor: | Glucocorticoid Modulatory Element Binding protein-1, ZINC ION | | Authors: | Surdo, P.L, Bottomley, M.J, Sattler, M, Scheffzek, K. | | Deposit date: | 2003-03-10 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and nuclear magnetic resonance analyses of the SAND domain from glucocorticoid modulatory element binding protein-1 reveals deoxyribonucleic acid and zinc binding regions
MOL.ENDOCRINOL., 17, 2003
|
|
2Y7S
 
 | |
2YPV
 
 | | Crystal structure of the Meningococcal vaccine antigen factor H binding protein in complex with a bactericidal antibody | | Descriptor: | 1,2-ETHANEDIOL, FAB 12C1, LIPOPROTEIN | | Authors: | Malito, E, Veggi, D, Bottomley, M.J. | | Deposit date: | 2012-11-01 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining a Protective Epitope on Factor H Binding Protein, a Key Meningococcal Virulence Factor and Vaccine Antigen.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
