1WJC
 
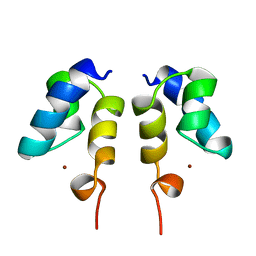 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (E FORM), NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJE
 
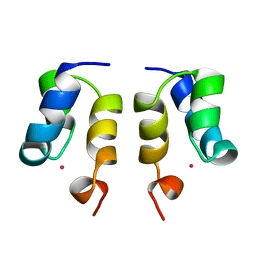 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CADMIUM ION, HIV-1 INTEGRASE | | Authors: | Cai, M, Gronenborn, A.M, Clore, G.M. | | Deposit date: | 1998-06-11 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
3ICS
 
 | |
2STT
 
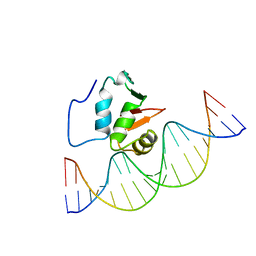 | | SOLUTION NMR STRUCTURE OF THE HUMAN ETS1/DNA COMPLEX, 25 STRUCTURES | | Descriptor: | DNA (5'-D(*TP*CP*GP*AP*AP*CP*TP*TP*CP*CP*GP*GP*CP*TP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*CP*CP*GP*GP*AP*AP*GP*TP*TP*CP*GP*A)-3'), ETS1 | | Authors: | Clore, G.M, Werner, M.H, Gronenborn, A.M. | | Deposit date: | 1996-08-05 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Correction of the NMR structure of the ETS1/DNA complex.
J.Biomol.NMR, 10, 1997
|
|
3ICR
 
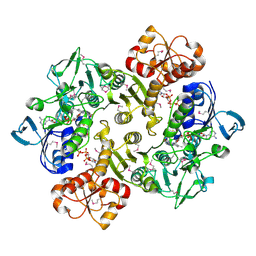 | | Crystal structure of oxidized Bacillus anthracis CoADR-RHD | | Descriptor: | COENZYME A, Coenzyme A-Disulfide Reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Wallen, J.R, Claiborne, A. | | Deposit date: | 2009-07-18 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking.
Biochemistry, 48, 2009
|
|
2STW
 
 | | SOLUTION NMR STRUCTURE OF THE HUMAN ETS1/DNA COMPLEX, RESTRAINED REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*TP*CP*GP*AP*AP*CP*TP*TP*CP*CP*GP*GP*CP*TP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*CP*CP*GP*GP*AP*AP*GP*TP*TP*CP*GP*A)-3'), ETS1 | | Authors: | Clore, G.M, Werner, M.H, Gronenborn, A.M. | | Deposit date: | 1996-08-05 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Correction of the NMR structure of the ETS1/DNA complex.
J.Biomol.NMR, 10, 1997
|
|
2MWH
 
 | | NMR solution structure of ligand-free OAA | | Descriptor: | Anti-HIV lectin OAA | | Authors: | Lee, D, Carneiro, M.G, Koharudin, L.M, Griesinger, C, Gronenborn, A.M, Ban, D, Sabo, T, Trigo-Mourino, P, Mazur, A. | | Deposit date: | 2014-11-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sampling of Glycan-Bound Conformers by the Anti-HIV Lectin Oscillatoria agardhii agglutinin in the Absence of Sugar.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6GAT
 
 | | SOLUTION NMR STRUCTURE OF THE L22V MUTANT DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13 BP DNA CONTAINING A TGATA SITE, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*AP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity.
J.Mol.Biol., 277, 1998
|
|
3HP8
 
 | | Crystal structure of a designed Cyanovirin-N homolog lectin; LKAMG, bound to sucrose | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Cyanovirin-N-like protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Koharudin, L.M.I, Furey, W, Gronenborn, A.M. | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A designed chimeric cyanovirin-N homolog lectin: Structure and molecular basis of sucrose binding.
Proteins, 77, 2009
|
|
3ICT
 
 | | Crystal structure of reduced Bacillus anthracis CoADR-RHD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COENZYME A, Coenzyme A-Disulfide Reductase, ... | | Authors: | Wallen, J.R, Claiborne, A. | | Deposit date: | 2009-07-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking.
Biochemistry, 48, 2009
|
|
2WFH
 
 | | The Human Slit 2 Dimerization Domain D4 | | Descriptor: | SLIT HOMOLOG 2 PROTEIN C-PRODUCT, SULFATE ION | | Authors: | Seiradake, E, von Philipsborn, A.C, Henry, M, Fritz, M, Lortat-Jacob, H, Jamin, M, Hemrika, W, Bastmeyer, M, Cusack, S, McCarthy, A.A. | | Deposit date: | 2009-04-06 | | Release date: | 2009-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and functional relevance of the Slit2 homodimerization domain.
EMBO Rep., 10, 2009
|
|
6HIR
 
 | |
2L2F
 
 | |
1WJA
 
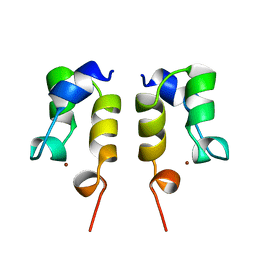 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJD
 
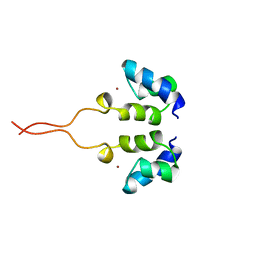 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (E FORM), NMR, 38 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
2P4V
 
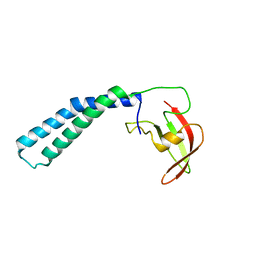 | | Crystal structure of the transcript cleavage factor, GreB at 2.6A resolution | | Descriptor: | Transcription elongation factor greB | | Authors: | Vassylyeva, M.N, Svetlov, V, Dearborn, A.D, Klyuyev, S, Artsimovitch, I, Vassylyev, D.G. | | Deposit date: | 2007-03-13 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The carboxy-terminal coiled-coil of the RNA polymerase beta'-subunit is the main binding site for Gre factors.
Embo Rep., 8, 2007
|
|
2EZX
 
 | |
2RGH
 
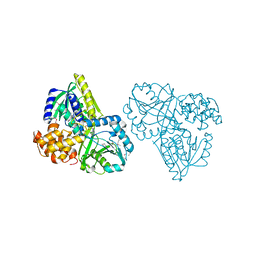 | | Structure of Alpha-Glycerophosphate Oxidase from Streptococcus sp.: A Template for the Mitochondrial Alpha-Glycerophosphate Dehydrogenase | | Descriptor: | Alpha-Glycerophosphate Oxidase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Colussi, T, Boles, W, Mallett, T.C, Karplus, P.A, Claiborne, A. | | Deposit date: | 2007-10-01 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of alpha-glycerophosphate oxidase from Streptococcus sp.: a template for the mitochondrial alpha-glycerophosphate dehydrogenase.
Biochemistry, 47, 2008
|
|
2RGO
 
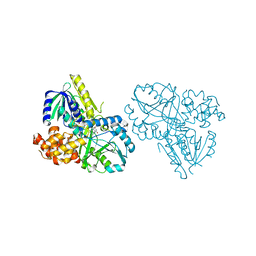 | | Structure of Alpha-Glycerophosphate Oxidase from Streptococcus sp.: A Template for the Mitochondrial Alpha-Glycerophosphate Dehydrogenase | | Descriptor: | Alpha-Glycerophosphate Oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Colussi, T, Boles, W, Mallett, T.C, Karplus, P.A, Claiborne, A. | | Deposit date: | 2007-10-04 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of alpha-glycerophosphate oxidase from Streptococcus sp.: a template for the mitochondrial alpha-glycerophosphate dehydrogenase.
Biochemistry, 47, 2008
|
|
2M65
 
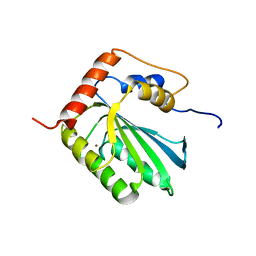 | | NMR structure of human restriction factor APOBEC3A | | Descriptor: | Probable DNA dC->dU-editing enzyme APOBEC-3A, ZINC ION | | Authors: | Byeon, I.L, Byeon, C, Ahn, J, Gronenborn, A.M. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity.
Nat Commun, 4, 2013
|
|
1BBN
 
 | |
1NER
 
 | |
1NEQ
 
 | |
2NBQ
 
 | |
1XS9
 
 | | A MODEL OF THE TERNARY COMPLEX FORMED BETWEEN MARA, THE ALPHA-CTD OF RNA POLYMERASE AND DNA | | Descriptor: | 5'-D(P*AP*TP*GP*CP*CP*AP*CP*GP*TP*TP*TP*TP*GP*CP*TP*AP*AP*AP*TP*C)-3', 5'-D(P*GP*AP*TP*TP*TP*AP*GP*CP*AP*AP*AP*AP*CP*GP*TP*GP*GP*CP*AP*T)-3', DNA-directed RNA polymerase alpha chain, ... | | Authors: | Dangi, B, Gronenborn, A.M, Rosner, J.L, Martin, R.G. | | Deposit date: | 2004-10-18 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Versatility of the carboxy-terminal domain of the alpha subunit of RNA polymerase in transcriptional activation: use of the DNA contact site as a protein contact site for MarA.
Mol.Microbiol., 54, 2004
|
|
